Page 271 - 2022_01-Haematologica-web
P. 271
Novel GFI1B variant dysregulates oncogenic factors
and III-1 carrying the c.648+5G>A variant, and similar to II- 4 who is wild-type (data not shown).
Expression level of alternative spliced products
The patient’s cDNA was used to clone the wild-type (330 amino acids [aa]; p37) and the exon 9 skipped (284 aa; p32) forms of GFI1B tagged to myc into an expression vector. Accordingly to what was observed during the RT-
A
B
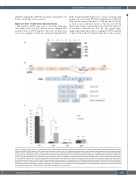
PCR on patients RNA (Figure 1C), colony screening using primers on exons 8 and 10 (Online Supplementary Table S1) detected the expected products of 246 bp and of 108 bp, as well as two additional bands of 312 bp and 174 bp (Figure 2A). Sanger sequencing showed that the addition- al products retained the last 66 bp (gggatcccggc- cgggtccagtcctgagcctgcacctgaccccccggggcctcatttcctccggcag) of intron 9 in both p37 and p32 forms due to the recogni-
C
Figure 2. GFI1B splicing isoforms. (A) Polymerase chain reaction (PCR) products obtained after amplification of a portion of GFI1B from different plasmids containing subcloned GFI1B cDNA (individual II-2) using primer aligning on exons 8 and 10. Fragments corresponding to the wild-type (246 basepairs [bp]) and skipped exon 9 (108 bp) forms of GFI1B are shown in lanes 1 and 4, and 2 and 5, respectively. Additional fragments of 312 bp (lane 7) and 174 bp (lanes 3 and 6) corresponding to p39 and p34, respectively, were also detected. M: molecular weight. (B) Schematic structure of the genomic GFI1B gene (gDNA) which includes the coding exons (exons 6-11; NM_004188.6). On the bottom, the representation of the full cDNA (p37) and the different isoforms identified. p32 is characterized by skipping of exon 9. p39 and p34 results in p37 and p32, respectively, which also retain the last 66 bp of intron 9 (of which the first and the last are listed in detail) due to recognition of a cryptic acceptor splice site “ag” (in bold). Arrows represent primers used for the amplification. (C) Differential expression level of the four GFI1B isoforms between patients (II-2 and III-1) and healthy controls (HC) perfomed by quantitative PCR using specific primers. Overall the wild-type isoform (p37) is the most represented in controls while p32 is significantly increased in patients (*P<0.05). Error bars represent the standard deviation of three independent experiments. Statistical analysis was performed using t-test. GDNA: genomic DNA; cDNA: coding DNA; aa: amino acids.
haematologica | 2022; 107(1)
263