Page 50 - 2021_09-Haematologica-web
P. 50
F. Barbaglio et al.
retrieved from the vessels after 3 days of co-culture and submitted to confocal analysis showed that GFP-tagged MEC1 cells populated the entire scaffold efficiently and homogeneously (Figure 1B). The model allows parallel analysis of CLL cells inside and outside the scaffold, reveal- ing that HS5 cells were needed in order to retain MEC1 cells
efficiently within the scaffold (Figure 1C).
Next, we used MEC1 cells genetically modified to down-
regulate HS1 expression (MEC1-HS1KD), already known to display increased BM homing capacity in vivo in a CLL xenograft model,9 and tested whether this could be repro- duced in our 3D ex-vivo model. We co-cultured in 3D either
A
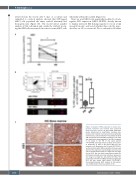
Figure 3. Analysis of HS1 expression in chronic lym- phocytic leukemia cells isolated from peripheral blood and bone marrow. (A) HS1 mRNA expression levels, determined by quantitative real-time poly- merase chain reaction in primary chronic lymphocytic leukemia (CLL) cells isolated from the peripheral blood (PB) and the bone marrow (BM) of patients (n=9). HS1 was significantly downregulated in the BM, compared to its levels in the PB of the same patient (*P=0.0498). (B) Image Stream analysis of intra-clon- al expression of HS1 in PB versus BM from CLL patients (n=4). By gating on the CLL pool (CD5+CD19+), we found mainly within BM a population that was neg- ative for HS1 expression (red rectangle and arrow indi- cate a representative image of a single CLL cell nega- tive for HS1). We also found a population positive for HS1 (black rectangle and arrow indicate a representa- tive image of a single CLL cell positive for HS1). The percentage of the PB and BM populations negative for HS1 are also shown (right panel) (*P=0.0187). Immunohistochemical (IHC) analysis of HS1 expres- sion in BM sections from CLL patients (n=4)
2338
haematologica | 2021; 106(9)
C
B