Page 225 - Haematologica May 2020
P. 225
Genomic alterations in high-risk CLL
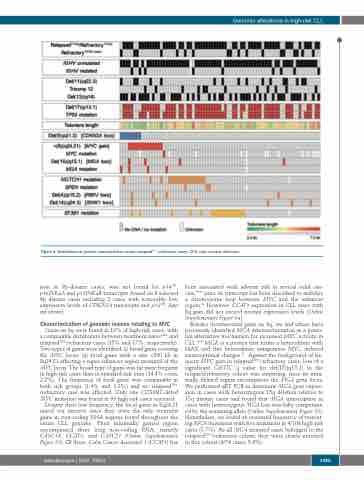
Figure 4. Distribution of genetic characteristics across relapsedTP53-/refractory cases. CNA: copy number alteration.
sion in 9p-disome cases, was not found for p14ARF, p16INK4A and p15INK4B transcripts (based on 8 selected 9p disome cases including 2 cases with noticeably low expression levels of CDKN2A transcripts and p14ARF; data not shown).
Characterization of genomic lesions relating to MYC Gains on 8q were found in 16% of high-risk cases, with a comparable distribution between treatment-naïveTP53- and relapsedTP53-/refractory cases (15% and 17%, respectively). Two types of gains were identified: (i) broad gains covering the MYC locus; (ii) focal gains with a size <500 kb in 8q24.21 affecting a super enhancer region proximal of the MYC locus. The broad type of gains was far more frequent in high-risk cases than in standard-risk ones (14.4% versus 2.2%). The frequency of focal gains was comparable in both risk groups (1.4% and 1.3%) and no relapsedTP53- /refractory case was affected. Only one COSMIC-listed
MYC mutation was found in 93 high-risk cases screened. Despite their low frequency, the focal gains in 8q24.21 raised our interest, since they were the only recurrent gains in non-coding DNA regions found throughout the entire CLL genome. Their minimally gained region encompassed three long non-coding RNA, namely CASC19, CCAT1, and CASC21 (Online Supplementary Figure S3). Of these, Colon Cancer Associated 1 (CCAT1) has
been associated with adverse risk in several solid can- cers,29,30 since its transcript has been described to stabilize a chromosome loop between MYC and the enhancer region.31 However, CCAT1 expression in CLL cases with 8q gain did not exceed normal expression levels (Online Supplementary Figure S4).
Besides chromosomal gains on 8q, we and others have previously identified MGA deletion/mutation as a poten- tial alternative mechanism for increased MYC activity in CLL.13,32 MGA is a protein that forms a heterodimer with MAX and this heterodimer antagonizes MYC induced transcriptional changes.33 Against the background of fre- quent MYC gain in relapsedTP53-/refractory cases, loss of a significant GISTIC q value for del(15)(q15.1) in the relapsed/refractory cohort was surprising, since its mini- mally deleted region encompasses the MGA gene locus. We performed qRT PCR to determine MGA gene expres- sion in cases with heterozygous 15q deletion relative to 15q disome cases and found that MGA transcription in cases with heterozygous MGA loss was fully compensat- ed by the remaining allele (Online Supplementary Figure S5). Nonetheless, we found an increased frequency of truncat- ing MGA mutations with five mutations in 4/108 high-risk cases (3.7%). As all MGA mutated cases belonged to the relapsedTP53-/refractory cohort, they were clearly enriched in this cohort (4/74 cases; 5.4%).
haematologica | 2020; 105(5)
1385