Page 227 - Haematologica May 2020
P. 227
Genomic alterations in high-risk CLL
AB
C
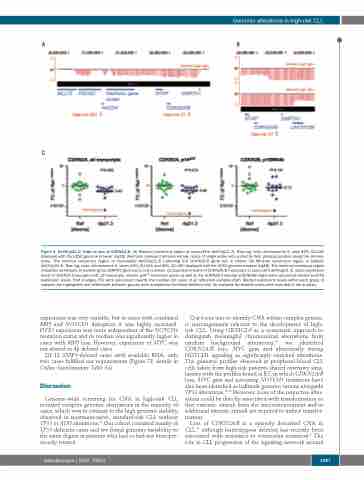
Figure 6. Del(9)(p21.3) leads to loss of CDKN2A/B. (A) Minimal consensus region of monoallelic del(9)(p21.3). Raw log2 ratio, chromosome 9, case #2O_CLL047 displayed with the UCSC genome browser (hg18). Red bars represent determined log2 ratios of single probe sets sorted by their physical position along the chromo- some. The minimal consensus region of monoallelic del(9)(p21.3) harboring the CDKN2A/B gene loci is shown. (B) Minimal consensus region of biallelic del(9)(p21.3). Raw log2 ratio, chromosome 9, cases #2O_CLL011 and #2O_CLL050 displayed with the UCSC genome browser (hg18). The minimal consensus region of biallelic del(9)(p21.3) harboring the DMRTA1 gene locus only is shwon. (C) Expression levels of CDKN2A/B transcripts in cases with del(9)(p21.3). Gene expression levels of CDKN2A transcripts (left: all transcripts, middle: p14ARF transcript alone) as well as the CDKN2B transcript p15INK4B (right) were calculated relative to ACTB expression levels. Fold changes (FC) were calculated towards the median DCt value of all reference samples (Ref). Median expression levels within each group of samples are highlighted and differences between groups were analyzed by the Mann-Whitney test. All available 9p-deleted cases were included in the analysis.
expression was very variable, but in cases with combined RBPJ and NOTCH1 disruption it was highly increased. DTX1 expression was more independent of the NOTCH1 mutation status and its median was significantly higher in cases with RBPJ loss. However, expression of MYC was not altered in 4p deleted cases.
Of 12 SNW1-deleted cases with available RNA, only two cases fulfilled our requirements (Figure 7F; details in Online Supplementary Table S4).
Discussion
Genome-wide screening for CNA in high-risk CLL revealed complex genomic aberrations in the majority of cases, which was in contrast to the high genomic stability observed in treatment-naïve, standard-risk CLL without TP53 or ATM alterations.13 Our cohort consisted mainly of TP53-deficient cases and we found genomic instability to the same degree in patients who had or had not been pre- viously treated.
Our focus was to identify CNA within complex genom- ic rearrangements relevant to the development of high- risk CLL. Using GISTIC2.0 as a systematic approach to distinguish meaningful chromosomal aberrations from random background alterations,16 we identified CDKN2A/B loss, MYC gain and abnormally strong NOTCH1 signaling as significantly enriched alterations. The genomic profiles observed in peripheral blood CLL cells taken from high-risk patients shared extensive simi- larities with the profiles found in RT, in which CDKN2A/B loss, MYC gain and activating NOTCH1 mutations have also been identified as hallmark genomic lesions alongside TP53 alterations.27,28 However, none of the respective alter- ations could be directly associated with transformation so that extrinsic stimuli from the microenvironment and/or additional intrinsic stimuli are required to induce transfor- mation.
Loss of CDKN2A/B is a sparsely described CNA in CLL,41 although homozygous deletion has recently been associated with resistance to venetoclax treatment.5 The role in CLL progression of the signaling network around
haematologica | 2020; 105(5)
1387