Page 226 - Haematologica May 2020
P. 226
J. Edelmann et al.
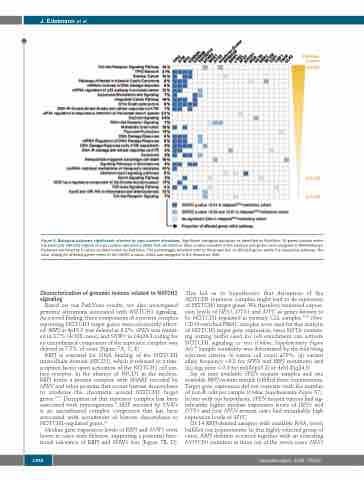
Figure 5. Biological pathways significantly affected by copy number alterations. Significant biological pathways as identified by PathVisio. All genes located within the minimally affected regions of copy number alterations (CNA) that are listed in Table 1 were included in the analysis and genes were assigned to WikiPathways. Pathways are listed by P values as determined via PathVisio. The percentages provided refer to the proportion of affected genes within the respective pathway. The color coding for affected genes refers to the GISTIC q value, which was assigned to the respective CNA.
Characterization of genomic lesions related to NOTCH1 signaling
Based on our PathVisio results, we also investigated genomic alterations associated with NOTCH1 signaling. As a novel finding, three components of a protein complex repressing NOTCH1 target genes were recurrently affect- ed: RBPJ in 4p15.2 was deleted in 8.2%; SPEN was mutat- ed in 3.7% (4/108 cases); and SNW1 in 14q24.3 coding for an unconfirmed component of the repressor complex was deleted in 7.5% of cases (Figure 7A, C, E).
RBPJ is essential for DNA binding of the NOTCH1 intracellular domain (NICD1), which is released as a tran- scription factor upon activation of the NOTCH1 cell sur- face receptor. In the absence of NICD1 in the nucleus, RBPJ forms a protein complex with SHARP encoded by SPEN and other proteins that recruit histone deacetylases to condense the chromatin around NOTCH1 target genes.34-36 Disruption of this repressor complex has been associated with tumorigenesis.37 SKIP encoded by SNW1 is an unconfirmed complex component that has been associated with recruitment of histone deacetylases to NOTCH1-regulated genes.34
Median gene expression levels of RBPJ and SNW1 were lower in cases with deletion, supporting a potential func- tional relevance of RBPJ and SNW1 loss (Figure 7B, D).
This led us to hypothesize that disruption of the NOTCH1 repressor complex might lead to de-repression of NOTCH1 target genes. We therefore measured expres- sion levels of HES1, DTX1 and MYC as genes known to be NOTCH1-regulated in primary CLL samples.38,39 Non- CD19-enriched PBMC samples were used for this analysis of NOTCH1 target gene expression, since EDTA-contain- ing sorting buffer used for cell enrichment can activate NOTCH1 signaling ex vivo (Online Supplementary Figure S6).40 Sample suitability was determined by the following selection criteria: (i) tumor cell count ≥70%; (ii) variant allele frequency >0.3 for SPEN and RBPJ mutations; and (iii) log2 ratio <-0.8 for del(4)(p15.2) or del(14)(q24.3)
Six of nine available SPEN mutant samples and one available RBPJ mutant sample fulfilled these requirements. Target gene expression did not correlate with the number of non-B cells per sample (Online Supplementary Figure S7). In line with our hypothesis, SPEN mutant tumors had sig- nificantly higher median expression levels of HES1 and DTX1 and four SPEN mutant cases had remarkably high expression levels of MYC.
Of 14 RBPJ-deleted samples with available RNA, seven fulfilled our requirements. In this highly selected group of cases, RBPJ deletion occurred together with an activating NOTCH1 mutation in three out of the seven cases. HES1
1386
haematologica | 2020; 105(5)