Page 223 - Haematologica May 2020
P. 223
Genomic alterations in high-risk CLL
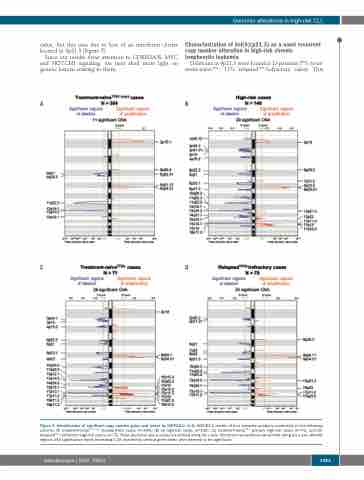
value, but this was due to loss of an interferon cluster located in 9p21.3 (Figure 5).
Since our results drew attention to CDKN2A/B, MYC and NOTCH1 signaling, we next shed more light on genetic lesions relating to them.
Characterization of del(9)(p21.3) as a novel recurrent copy number alteration in high-risk chronic lymphocytic leukemia
Deletionsin9p21.3werefoundin13patients(7%treat- ment-naïveTP53-; 11% relapsedTP53-/refractory cases). This
AB
CD
Figure 3. Identification of significant copy number gains and losses by GISTIC2.0. (A-D) GISTIC2.0 results of four separate analyses conducted on the following cohorts: (A) treatment-naïveTP53 intact standard-risk cases (n=304); (B) all high-risk cases (n=146); (C) treatment-naïveTP53- primary high-risk cases (n=71); and (D) relapsedTP53-/refractory high-risk cases (n=75). False discovery rate q values are plotted along the x axis. Chromosomal positions are plotted along the y axis. Altered regions with significance levels exceeding 0.25 (marked by vertical green lines) were deemed to be significant.
haematologica | 2020; 105(5)
1383