Page 221 - Haematologica May 2020
P. 221
Genomic alterations in high-risk CLL
Whitney tests. All statistical tests were two-sided and conducted in GraphPad Prism® version 7.0.
Further details are available in the Online Supplementary Methods. Results
Cohort characteristics
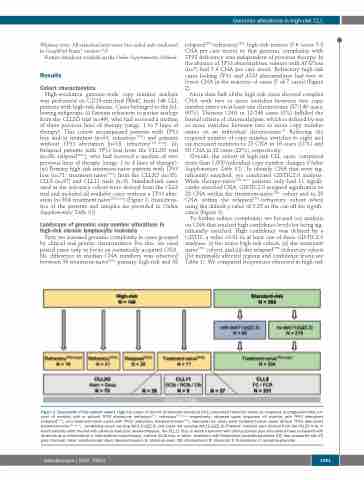
High-resolution genome-wide copy number analysis was performed on CD19-enriched PBMC from 146 CLL patients with high-risk disease. Cases belonged to the fol- lowing subgroups. (i) Patients refractory to purine analogs from the CLL2O trial (n=49), who had received a median of three previous lines of therapy (range, 1 to 7 lines of therapy). This cohort encompassed patients with TP53 loss and/or mutation (n=31; refractoryTP53-) and patients without TP53 aberration (n=18; refractoryTP53 intact). (ii) Relapsed patients with TP53 loss from the CLL2O trial (n=26; relapsedTP53-), who had received a median of two previous lines of therapy (range, 1 to 4 lines of therapy). (iii) Primary high-risk treatment-naïve patients with TP53 loss (n=71; treatment-naïveTP53-) from the CLL2O (n=35), CLL8 (n=27) and CLL11 trials (n=9). Standard-risk cases used as the reference cohort were derived from the CLL8 trial and included all available cases without a TP53 alter- ation (n=304; treatment naïveTP53 intact) (Figure 1; characteris- tics of the patients and samples are provided in Online Supplementary Table S1).
Landscape of genomic copy number alterations in high-risk chronic lymphocytic leukemia
First, we assessed genomic complexity in cases grouped by clinical and genetic characteristics. For this, we used paired cases only to focus on somatically acquired CNA. No difference in median CNA numbers was observed between 39 treatment-naïveTP53- primary high-risk and 38
relapsedTP53-/refractoryTP53- high-risk tumors (5.6 versus 5.8 CNA per case mean) so that genomic complexity with TP53 deficiency was independent of previous therapy. In the absence of TP53 abnormalities, tumors with ATM loss (n=7) had 5.4 CNA per case mean. Refractory high-risk cases lacking TP53 and ATM abnormalities had two or fewer CNA in the majority of cases (5 of 7 cases) (Figure 2).
More than half of the high-risk cases showed complex CNA with two or more switches between two copy number states on at least one chromosome (87/146 cases, 60%). Thirteen CNA in 12/146 cases (8%) fulfilled the formal criteria of chromothripsis, which is defined by ten or more switches between two or more copy number states on an individual chromosome.26 Reducing the required number of copy number switches to eight and six increased numbers to 23 CNA in 16 cases (11%) and 50 CNA in 32 cases (22%), respectively.
Overall, the cohort of high-risk CLL cases comprised more than 1,500 individual copy number changes (Online Supplementary Table S2). To identify CNA that were sig- nificantly enriched, we conducted GISTIC2.0 analysis. While therapy-naïveTP53 intact patients only had 11 signifi- cantly enriched CNA, GISTIC2.0 assigned significance to 28 CNA within the treatment-naïveTP53- cohort and to 20 CNA within the relapsedTP53-/refractory cohort when using the default q value of 0.25 as the cut-off for signifi- cance (Figure 3).
To further reduce complexity, we focused our analysis on CNA that reached high confidence levels for being sig- nificantly enriched. High confidence was defined by a GISTIC q value <0.01 in at least one of three GISTIC2.0 analyses: (i) the entire high-risk cohort, (ii) the treatment naïveTP53- cohort, and (iii) the relapsedTP53-/refractory cohort (for minimally affected regions and confidence levels see Table 1). We compared frequencies observed in high-risk
Figure 1. Description of the sample cohort. High-risk cases of chronic lymphocytic leukemia (CLL) comprised refractory cases (no response or progression-free sur- vival <6 months) with or without TP53 alterations (refractoryTP53-, refractoryTP53 intact, respectively), relapsed cases (response >6 months) with TP53 alterations (relapsedTP53-), and treatment-naïve cases with TP53 alterations (treatment-naïveTP53-). Standard-risk cases were treatment-naïve cases without TP53 alterations (treatment-naïveTP53 intact), comprising cases carrying del(11)(q22.3) and cases not carrying del(11)(q22.3). Patients’ samples were derived from the CLL2O trial, in which patients were treated with alemtuzumab plus dexamethasone, the CLL11 trial, in which treatment with obinutuzumab plus chlorambucil was compared with rituximab plus chlorambucil or chlorambucil monotherapy, and the CLL8 trial, in which treatment with fludarabine/cyclophosphamide (FC) was compared with FC plus rituximab. Alem: alemtuzumab; dexa: dexamethasone; G: obinutuzumab; Clb: chlorambucil; R: rituximab; F: fludarabine; C: cyclophosphamide
haematologica | 2020; 105(5)
1381