Page 218 - Haematologica April 2020
P. 218
A. Mikulasova et al.
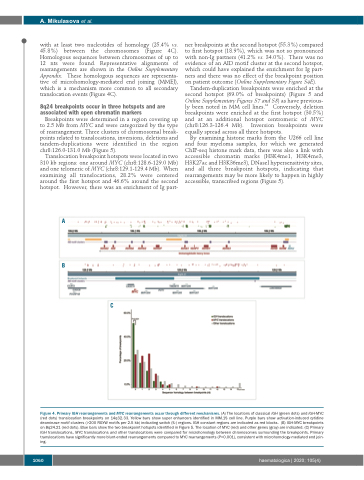
with at least two nucleotides of homology (25.4% vs. 45.8%) between the chromosomes (Figure 4C). Homologous sequences between chromosomes of up to 12 nts were found. Representative alignments of rearrangements are shown in the Online Supplementary Appendix. These homologous sequences are representa- tive of microhomology-mediated end joining (MMEJ), which is a mechanism more common to all secondary translocation events (Figure 4C).
8q24 breakpoints occur in three hotspots and are associated with open chromatin markers
Breakpoints were determined in a region covering up to 2.5 Mb from MYC and were categorized by the type of rearrangement. Three clusters of chromosomal break- points related to translocations, inversions, deletions and tandem-duplications were identified in the region chr8:126.0-131.0 Mb (Figure 5).
Translocation breakpoint hotspots were located in two 310 kb regions: one around MYC (chr8:128.6-129.0 Mb) and one telomeric of MYC (chr8:129.1-129.4 Mb). When examining all translocations, 28.2% were centered around the first hotspot and 46.6% around the second hotspot. However, there was an enrichment of Ig part-
ner breakpoints at the second hotspot (55.3%) compared to first hotspot (18.9%), which was not so pronounced with non-Ig partners (41.2% vs. 34.0%). There was no evidence of an AID motif cluster at the second hotspot, which could have explained the enrichment for Ig part- ners and there was no effect of the breakpoint position on patient outcome (Online Supplementary Figure S4E).
Tandem-duplication breakpoints were enriched at the second hotspot (69.0% of breakpoints) (Figure 5 and Online Supplementary Figures S7 and S8) as have previous- ly been noted in MM cell lines.34 Conversely, deletion breakpoints were enriched at the first hotspot (30.5%) and at an additional hotspot centromeric of MYC (chr8:126.3-126.4 Mb). Inversion breakpoints were equally spread across all three hotspots.
By examining histone marks from the U266 cell line and four myeloma samples, for which we generated ChIP-seq histone mark data, there was also a link with accessible chromatin marks (H3K4me1, H3K4me3, H3K27ac and H3K36me3), DNaseI hypersensitivity sites, and all three breakpoint hotspots, indicating that rearrangements may be more likely to happen in highly accessible, transcribed regions (Figure 5).
A
B
C
Figure 4. Primary IGH rearrangements and MYC rearrangements occur through different mechanisms. (A) The locations of classical IGH (green dots) and IGH-MYC (red dots) translocation breakpoints on 14q32.33. Yellow bars show super enhancers identified in MM.1S cell line. Purple bars show activation-induced cytidine deaminase motif clusters (>200 RGYW motifs per 2.5 kb) indicating switch (S-) regions. IGH constant regions are indicated as red blocks. (B) IGH-MYC breakpoints on 8q24.21 (red dots). Blue bars show the two breakpoint hotspots identified in Figure 5. The location of MYC (red) and other genes (gray) are indicated. (C) Primary IGH translocations, MYC translocations and other translocations were compared for microhomology between chromosomes surrounding the breakpoints. Primary translocations have significantly more blunt-ended rearrangements compared to MYC rearrangements (P<0.001), consistent with microhomology-mediated end join- ing.
1060
haematologica | 2020; 105(4)