Page 57 - Haematologica Vol. 107 - September 2022
P. 57
ARTICLE - Genetic abnormalities in older ALL patients T. Creasey et al.
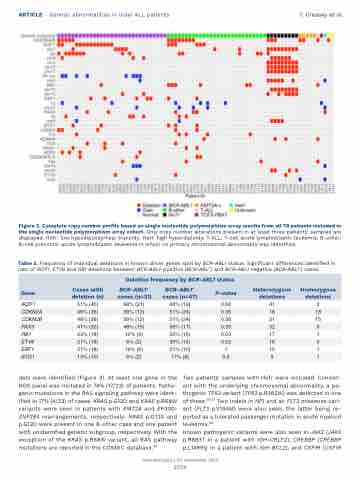
Figure 2. Complete copy number profile based on single nucleotide polymorphism array results from all 78 patients included in the single nucleotide polymorphism array cohort. Only copy number alterations present in at least three patients' samples are displayed. HoTr: low hypodiploidy/near triploidy; HeH: high hyperdiploidy; T-ALL: T-cell acute lymphoblastic leukemia; B-other: B-cell precursor acute lymphoblastic leukemia in which no primary chromosomal abnormality was identified.
Table 2. Frequency of individual deletions in known driver genes split by BCR-ABL1 status. Significant differences identified in rate of IKZF1, ETV6 and RB1 deletions between BCR-ABL1-positive (BCR-ABL+) and BCR-ABL1 negative (BCR-ABL1-) cases.
Deletion frequency by BCR-ABL1 status
Gene
Cases with deletion (n)
BCR-ABL1+
cases (n=31)
BCR-ABL1–
cases (n=47)
P-value
Heterozygous deletions
Homozygous deletions
IKZF1
51% (40)
68% (21)
40% (19)
0.02
41
2
CDKN2A
46% (36)
39% (12)
51% (24)
0.36
18
18
CDKN2B
46% (36)
39% (12)
51% (24)
0.36
21
15
PAX5
41% (32)
48% (15)
36% (17)
0.35
32
0
RB1
23% (18)
10% (3)
32% (15)
0.03
17
1
ETV6
21% (16)
6% (2)
30% (14)
0.02
16
0
EBF1
21% (16)
19% (6)
21% (10)
1
15
1
BTG1
13% (10)
6% (2)
17% (8)
0.3
9
1
dels were identified (Figure 3). At least one gene in the NGS panel was mutated in 74% (17/23) of patients. Patho- genic mutations in the RAS signaling pathway were ident- ified in 17% (4/23) of cases. KRAS p.G12D and KRAS p.R68W variants were seen in patients with KMT2A and EP300- ZNF384 rearrangements, respectively. NRAS p.G12S and p.G12D were present in one B-other case and one patient with unidentified genetic subgroup, respectively. With the exception of the KRAS p.R68W variant, all RAS pathway mutations are reported in the COSMIC database.25
Two patients’ samples with HoTr were included. Consist- ent with the underlying chromosomal abnormality, a pa- thogenic TP53 variant (TP53 p.R282W) was detected in one of these.26,27 Two indels in NF1 and an FLT3 missense vari- ant (FLT3 p.V194M) were also seen, the latter being re- ported as a tolerated passenger mutation in acute myeloid leukemia.28
Known pathogenic variants were also seen in JAK2 (JAK2 p.R683T in a patient with IGH-CRLF2), CREBBP (CREBBP p.L1499Q in a patient with IGH-BCL2), and CSF1R (CSF1R
Haematologica | 107 September 2022
2056