Page 56 - Haematologica Vol. 107 - September 2022
P. 56
ARTICLE - Genetic abnormalities in older ALL patients AB
T. Creasey et al.
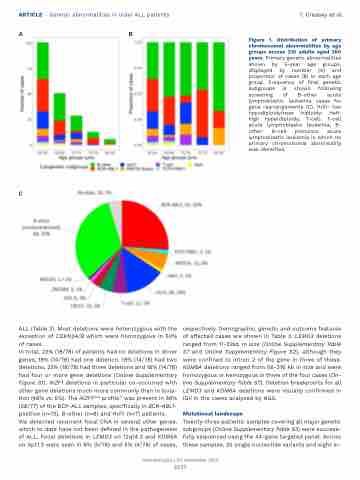
Figure 1. Distribution of primary chromosomal abnormalities by age groups across 210 adults aged ≥60 years. Primary genetic abnormalities shown by 5-year age groups, displayed by number (A) and proportion of cases (B) in each age group. Frequency of final genetic subgroups is shown following screening of B-other acute lymphoblastic leukemia cases for gene rearrangements (C). HoTr: low hypodiploidy/near triploidy; HeH: high hyperdiploidy; T-cell: T-cell acute lymphoblastic leukemia; B- other: B-cell precursor acute lymphoblastic leukemia in which no primary chromosomal abnormality was identified.
C
ALL (Table 2). Most deletions were heterozygous with the exception of CDKN2A/B which were homozygous in 50% of cases.
In total, 23% (18/78) of patients had no deletions in driver genes, 18% (14/78) had one deletion, 18% (14/78) had two deletions, 23% (18/78) had three deletions and 18% (14/78) had four or more gene deletions (Online Supplementary Figure S1). IKZF1 deletions in particular co-occurred with other gene deletions much more commonly than in isola- tion (46% vs. 5%). The IKZF1plus profile17 was present in 36% (28/77) of the BCP-ALL samples, specifically in BCR-ABL1- positive (n=13), B-other (n=8) and HoTr (n=7) patients.
We detected recurrent focal CNA in several other genes, which to date have not been defined in the pathogenesis of ALL. Focal deletions in LEMD3 on 12q14.3 and KDM6A on Xp11.3 were seen in 6% (5/78) and 5% (4/78) of cases,
respectively. Demographic, genetic and outcome features of affected cases are shown in Table 3. LEMD3 deletions ranged from 11-32kb in size (Online Supplementary Table S7 and Online Supplementary Figure S2), although they were confined to intron 2 of the gene in three of these. KDM6A deletions ranged from 56-316 kb in size and were homozygous or hemizygous in three of the four cases (On- line Supplementary Table S7). Deletion breakpoints for all LEMD3 and KDM6A deletions were visually confirmed in IGV in the cases analyzed by NGS.
Mutational landscape
Twenty-three patients’ samples covering all major genetic subgroups (Online Supplementary Table S3) were success- fully sequenced using the 44-gene targeted panel. Across these samples, 25 single nucleotide variants and eight in-
Haematologica | 107 September 2022
2055