Page 94 - 2021_03-Haematologica-web
P. 94
M.A. Bauer et al.
AB
CD
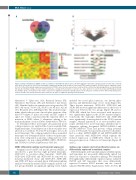
Figure 3. Hotspot mutations in SF3B1 results in a number of alternatively spliced genes. (A) Venn diagram of alternative splicing results from the three methods used. (B) A volcano plot of delta-PSI in relationship to significant splicing events between control and SF3B1 mutated samples. Red dots indicate significant events (P≤0.05). Differential splicing analysis was performed by SUPPA2. Genes that are labeled were found by all three differential splicing algorithms as statistically sig- nificant after multiple testing correction. Delta-PSI indicates a change in the inclusion or exclusion of a splice junction or exon in the mutated SF3B1 group versus the wild-type group. (C) Heatmap of the relative expression of significant alternative splicing features for the 18 genes found by all three differential splicing tools. Features include exon, known and novel splice junctions. (D) Types of significant splicing events identified.
Alternative 3’ Splice-sites (A3), Retained Introns (RI), Alternative First Exons (AF) and Alternative Last Exons (AL). Identified splice site variants were categorized as 418 (60%) AF exons, 76 (9%) SE, 55 (8%) A3, 45 (6%) A5, 26 (4%) RI and 12 (2%) MX (Figure 3D). The Leafcutter algo- rithm, which takes account of novel splice junctions, iden- tified 373 cryptic 5’ splice site events and 527 cryptic 3’ splice site events consistent with the expected effect of mutation of SF3B1 where 3’ alternative splicing is the main mechanism of action (Online Supplementary Table S5).
To determine if these novel splice sites generated novel coding sequences, we analyzed the de novo assembled full- length transcripts. Visual inspection of the novel splice sites allowed us to attribute 19 of the 39 novel splice sites to 20 novel transcripts. The coding potential for these assembled transcripts was determined and 16 of the novel splice junc- tions were categorized as protein coding and four were determined to be non-coding (CREBZF, MPC1, PKHD1L1, and TXNDC11) (Online Supplementary Table S6).
MZB1 differential splicing and transcript expression Marginal zone B and B1 cell specific protein (MZB1), is encoded by a gene that has eight potential transcripts. MZB1-201 is protein-coding transcript, MZB1-202, MZB1-204, MZB1-205, and MZB1-208 are removed via nonsense-medicated decay and MZB1-203, MZB1-206, and MZB1-207 have a retained intron. Differential splicing analysis between the mutated SF3B1 samples and the con- trol group identified six significant splicing events which
included two novel splice junctions, one known splice junction, and differential usage of two exons (Figure 4A). Three known transcripts, MZB1-203, MZB1-204 and MZB1-205 showed significant differences in the levels of expression between the two groups (Figure 4B and C). MZB1-205, which has been associated with apoptosis, was significantly higher in the SF3B1 mutant samples. Conversely, the transcripts MZB1-203 and MZB1-204 were significantly down-regulated in the SF3B1 mutant samples. Manual inspection of de novo-assembled tran- scripts identified two novel transcripts identified due to alternative splicing; designated MSTRG.24254.14 and MSTRG.24253.16. They had protein coding potential scores of 0.96 and 0.71 respectively and were both identi- fied as coding. MSTRG.24253.14 and MSTRG.24253.16 share junctions with MZB1-204, which explains the reduced expression of MZB1-204, which is due to increased expression of these novel transcripts (Figure 4B and C).
Spliceosome complex and cell proliferation genes are differentially expressed in mutated samples
Differential gene and transcript analysis identified genes and transcripts not necessarily differentially spliced but altered in response to alternative splicing. We identified 234 significantly differentially expressed genes (adjusted P≤0.05) and 365 transcripts that were differentially expressed between SF3B1 mutants and the controls (adjusted P≤0.05) (Online Supplementary Tables S7-S10). Of
740
haematologica | 2021; 106(3)