Page 222 - Haematologica April 2020
P. 222
A. Mikulasova et al.
duplications, another with non-Ig translocations and dele- tions, and the third with deletions and inversions. All breakpoints surrounding MYC result in increased expres- sion of the oncogene, but inter-chromosomal transloca- tions result in the largest increase in expression.
In this dataset, we have used 1,267 NDMM patient samples (of which 36.0% had MYC abnormalities) using next generation sequencing consisting of whole genome, exome and targeted panel data. The frequency of MYC abnormalities reported here is higher than previously seen using other techniques, such as karyotyping or fluores- cence in situ hybridization (FISH). This is likely due to the increased resolution of sequencing technologies that can identify small insertions or deletions as well as transloca- tions involving infrequent partner chromosomes. In addi- tion, the complexity of breakpoints at 8q24 makes the placement of FISH probes difficult if all abnormalities are to be detected. The scale of this analysis has allowed us to define the molecular breakpoints surrounding MYC with unparalleled accuracy and without technical bias. One of the two rearrangement hotspots involved in inter-chromo- somal translocations in MM is also seen in other B-cell malignancies. In Burkitt’s lymphoma, two breakpoint clusters within exon 1 and intron 1 of MYC were defined, which corresponds in location to the non-Ig rearrange- ment hotspot in MM.26 The same cluster is seen in diffuse large B-cell lymphoma, where other random breakpoints are also seen scattered both centromeric and telomeric of
MYC.25 Both of these studies looked at relatively small numbers of samples (78 and 17, respectively) and used older techniques, such as long distance PCR and FISH, to detect the breakpoints. It may be that there are also other breakpoint hotspots similar to MM in other B-cell malig- nancies.
The main chromosomal partner to MYC through inter- chromosomal rearrangements is chromosome 14, specifi- cally the IGH locus. In Burkitt’s lymphoma, the IGH-MYC breakpoints on this chromosome lie almost exclusively within the switch regions (87%), upstream of the IGH constant regions.26 The remaining 13% are within the joining region of the locus. These breakpoints are consis- tent with the IGH-MYC rearrangement, being a primary event in Burkitt’s lymphoma, occurring in 70-80% of patients.40 In contrast, in MM, we clearly see that IGH- MYC breakpoints within the IGH locus are not in the switch or joining regions; instead, they are spread out across the constant regions of the locus. This spread is dis- tinct from the five common primary translocation break- points in MM [t(4;14), t(11;14), etc.] which are restricted to the switch and joining regions. Even those with MYC breakpoints within switch regions (6.9% of IGH-MYC rearrangements) also have primary rearrangements or are hyperdiploid. This indicates that the IGH-MYC rearrange- ments are secondary events in MM and probably occur through a different molecular mechanism to the primary translocation events. It is known that the primary translo-
AB
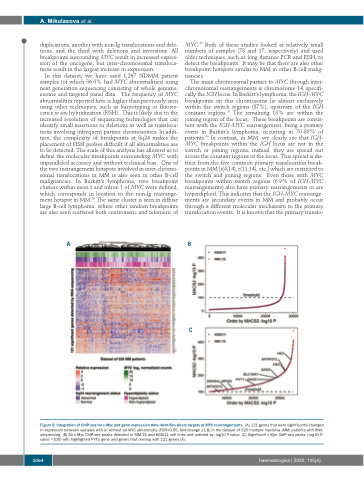
Figure 8. Integration of ChIP-seq for c-Myc and gene expression data identifies direct targets of MYC rearrangements. (A) 121 genes that were significantly changed in expression between samples with or without an MYC abnormality (FDR<0.05, fold-change ≥1.8) in the dataset of 526 multiple myeloma (MM) patients with RNA- sequencing. (B) All c-Myc ChIP-seq peaks detected in MM.1S and KMS11 cell lines and ordered by -log10 P-value. (C) Significant c-Myc ChIP-seq peaks (-log10 P- value >100) with highlighted PVT1 gene and genes that overlap with 121 genes (A).
C
1064
haematologica | 2020; 105(4)