Page 221 - Haematologica April 2020
P. 221
MMEJ drives 8q24 rearrangements in myeloma
binding sites in two MM cell lines, MM.1S and KMS11, both of which have an MYC rearrangement. The peaks with a significance P<10-100 using MACS2 in either cell line were considered significant and accounted for 4.7% of peaks (1,266 of 27,006) (Figure 8B). The peaks were com- pared to the 121 genes that were significantly changed in expression (Figure 8A). Six genes were in the intersection between over-expressed and significant peaks: HK2, MTHFD1L, SLC19A1, MFNG, SNHG4, GAS5, (Figure 8C). Using less stringent ≥1.3 fold-change cut-off that pro- vided 1,801 genes, of which 40.8% (735 of 1,801) were over-expressed, the intersection of over-expressed genes and those with a significant MYC binding peak was 25.3% (186 of 735). At the top of the list of 186 genes ordered by ChIP-seq -log10 P, we detected upregulation of the genes with known or potential oncogenic activity such as genes promoting cell proliferation, tumor growth and/or inhibition of apoptosis (SNHG15, PPAN, MAT2A, METAP1D, MTHFD2, SNHG17), translation factors (EIF3B, EIF4A1, EEF1B2), and genes involved in ribosome biosynthesis (RPL10A, RPL35, RPL23A, RPSA, RPL13, WDR43).
Importantly, we identified HK2 and PVT1 as direct tar- gets of MYC. HK2 is one of the most significant genes detected by ChIP-seq in both cell lines (-log10 P>200) (Figure 8C), as well as having the highest fold-change using RNA-sequencing analysis (Online Supplementary Table S7). This gene is an interesting direct target of MYC as it is part of the glucose metabolism pathway and would lead to increased energy metabolism and proliferation. PVT1 showed a smaller fold-change by RNA-sequencing analysis (approx. 1.4) but had a significant c-Myc protein binding site identified by ChIP-seq, meaning that overex- pression of PVT1 is likely to be a downstream effect of MYC overexpression. This leads to a positive feedback loop and even higher MYC expression, as PVT1 positively regulates MYC expression.39
Discussion
We show that MYC breakpoints in myeloma are clus- tered in three main hotspots on chromosome 8, one of which is associated with Ig translocations and tandem-
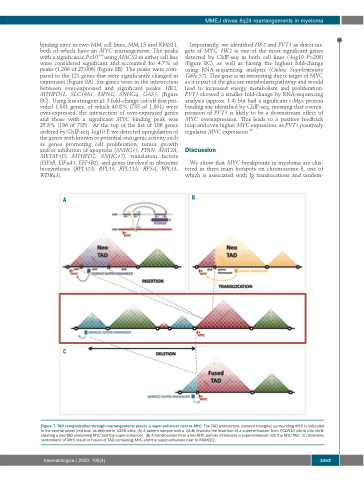
A
B
C
Figure 7. TAD reorganization through rearrangements places a super-enhancer next to MYC. The TAD architecture (colored triangles) surrounding MYC is indicated in the central panel (red box) as defined in U266 cells. (A) A patient sample with a t(4;8) involves the insertion of a super-enhancer from PCDH10 (chr4) into chr8, creating a neo-TAD containing MYC and the super-enhancer. (B) A translocation from a key MYC partner introduces a super-enhancer into the MYC TAD. (C) Deletions centromeric of MYC result in fusion of TAD containing MYC and the super-enhancer next to NSMCE2.
haematologica | 2020; 105(4)
1063