Page 246 - Haematologica March 2020
P. 246
B. Federmann et al.
assay and compared it with IHC and RT-qPCR. Additionally, the TP53 status was investigated and corre- lated with SOX11 mRNA levels. We show that RNAscope ISH assay is a reliable method to quantitate SOX11 mRNA levels within the histopathological context. Importantly, and in overall agreement with the RT-qPCR results, we demonstrated that SOX11 mRNA levels show a very broad range in cMCL and confirmed the negative or low mRNA levels in nnMCL cases. An interesting finding of this study was the correlation of TP53 mutations with low/negative SOX11 mRNA scores in all investigated sub- types of MCL (P=0.0007).
RNAscope is a relatively new ISH technology with a probe design strategy that allows the visualization of RNA expression in the context of preserved tissue morpholo- gy.33,39 In recent years, several studies confirmed the suit- ability of this method for the quantification of mRNA lev- els in FFPE tissues; however, SOX11 has not been investi- gated.34,40,41 The diagnostic importance of SOX11 expres- sion for identifying specific subsets of MCL has been con- firmed in many studies; however, its relevance as a prog- nostic marker is controversial. One possible explanation is that IHC, as a predominantly used technique, is not a quantitative method and does not provide reliable infor- mation about SOX11 expression levels. The current gold standard for quantitative gene expression analysis is RT- qPCR.42 However, this method lacks the morphological context, and in our study, a wide range of SOX11 mRNA level was found that not always correlated with the RNAscope assay. Similar findings were described by Lord et al., who also found a wide range of SOX11 mRNA lev-
els by RT-qPCR and were not able to define a natural cut- off that could stratify cases with low protein expression by IHC.43 The highly variable SOX11 expression in MCL could indicate that not only the presence or absence of the protein, but also its level may be important for the behav- ior of the malignant cells. Nevertheless, a similar number of cases, with high expression of SOX11 mRNA, was found in the group with blastoid morphology when com- pared with classic morphology (62% vs. 59%).
The oncogenic potential of SOX11 has been extensively studied in the last years.16,22-24,27,44,45 SOX11 has been shown to promote tumor growth via the induction of angiogene- sis by regulating PDGFA.23 Recently, it has also been described that SOX11 binds and transcriptionally regu- lates two genes important for the modulation of microen- vironment-tumor interactions, CXCR4 (C-X-C motif chemokine receptor 4) and PTK2, encoding for focal adhe- sion kinase (FAK).44 These genes are essential for tumor cell migration and adhesion to stromal cells in the bone mar- row.46 Furthermore, SOX11 has been described to corre- late also with high proliferation activity and aggressive behaviour of the disease.22,47,48 The accurate quantification of SOX11 expression opens the possibility to investigate the potential influence of SOX11 levels, and thus, might help to refine both the biological functions in MCL and to determine whether it can be used as a prognostic marker. The correlation between RNAscope, IHC and RT-qPCR was in general good; however, the automated RNAscope technology, direct visualization in tissue specimens and easy quantification of the signals makes this technology very attractive for diagnosis and research purposes. The
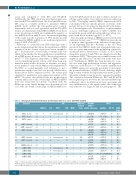
Table 3. Histological, immunohistochemical and molecular data of 13 cases with TP53 mutation.
Case #
6
Diagnosis
cMCL - classic
IHC
D1 P53 % CD5
60 heterogenous 35
3 + 10 80 + 80
70 + 30 30 + 80 80 + 80
90 heterogenous 80
90 + 90
90-20 80 heterogenous NA 90 heterogenous 5 - + 90 90+80
RNAscope RT-qPCR
TP53
mutation VAF [%] CADD Score
c.743G>A 77 27.3
SOX11 %
<10 +
>10 + <10 +
>10 + <10 + >10 -
<10 +
>10 +
<10 + 0 + 0 + 0 + 0 +
MIB %
1
4 1
SOX11
Score
10
SOX11
relative expression
p.R248Q
42 cMCL-classic 64 cMCL-classic
25 cMCL-blastoid 34 cMCL - blastoid * 37 cMCL-blastoid
58 cMCL - blastoid
66 cMCL-blastoid
80 p.L201Afs c.601_602del 12 19.2
1 p.R248W c.742C>T p.P278R c.833C>G
7 26.8 8 26.4
38 28.6
99 24.1
13 25.7 6 22.3 8 25.3
70 25.3
30 28.6
p.Y220C c.659A>G
p.V218G c.653T>G
p.Y205N c.613T>A
p.V272G c.815T>G
p.R273* c.815_816insTTGAGGT29 NA
21 p.P278S c.832C>T 5 p.R175H c.524G>A
3
1
4 NA p.R282W c.844C>T
p.R158H c.473G>A
p.R273C c.817C>T
1 NA p.R273C c.817C>T
3 NA p.P278S c.832C>T
9 15 16 23 35
nnMCL (spleen) nnMCL (BM) nnMCL (BM) nnMCL (BM) nnMCL (BM)
1 0
0 1
0 0
0 0
58 29.5 77 30 35 24.2 58 23.4
0 1 p.S127F c.380C>T 16 27
p.R248W c.742C>T 7 26.8
cMCL: conventional mantle cell lymphoma; D1-MCL: cyclinD1 negative mantle cell lymphoma; nnMCL: non-nodal leukemic mantle cell lymphoma; IHC: immunohistochemistry; RT-,PCR: real-time quantitative PCR; NGS: next generation sequencing; VAF: variant allelic frequency; NA: not available; * diagnosis of MCL made in pleura effusion; mutations with a CADD algorithm score >15 are considered deleterious.
760
haematologica | 2020; 105(3)