Page 126 - Haematologica May 2022
P. 126
S. Napoli et al.
B
C
D
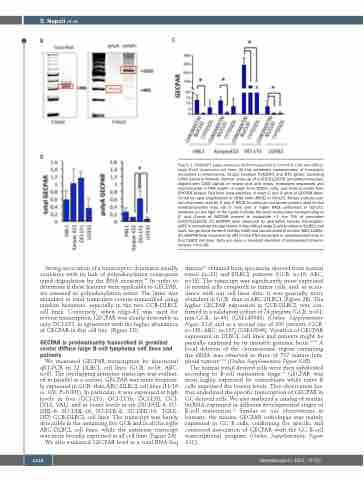
Figure 1. POU2AF1 super-enhancer derived transcript in normal B cells and diffuse large B-cell lymphoma cell lines. (A) Top: schematic representation of transcripts annotated in chromosome 11q23, between POU2AF1 and BTG genes, according UCSC Genome Browser. Bottom: close-up of LOC100132078 annotated transcript, aligned with CAGE signals on strand plus and minus, transcripts sequenced and reconstructed in RNA polyA+ or polyA- from CD20+ cells, and histone marks from ENCODE project. Red lines show positions of exact 5' and 3' ends of GECPAR deter- mined by rapid amplification of cDNA ends (RACE) in OCI-LY1. Arrows indicate posi- tion of primers used for 5' and 3' RACE, in particular red arrows primers used for the retrotranscription step. (B) 5’ (left) and 3’ (right) RACE performed in OCI-LY1. Numbers on the right of the bands indicate the exact nucleotides corresponding to 5’ and 3’ends of GECPAR respect to nucleotide +1, the TSS of annotated LOC100132078. (C) GECPAR level measured by quatitative reverse transcription (qRT) in subcellular compartments in four diffuse large B-cell lymphoma (DLBCL) cell lines, two germinal center B cell-like (GCB) and two activated B cell-like (ABC)-DLBCL. (D) GECPAR level measured by qRT in total RNA transcripts or polyadenylated only, in four DLBCL cell lines. Data are mean ± standard deviation of independent determi- nations. *P<0.05.
Strong association of a transcript to chromatin usually correlates with its lack of polyadenylation consequent rapid degradation by the RNA exosome.32 In order to determine if these features were applicable to GECPAR, we assessed its polyadenylation status. The latter was abundant in total transcripts reverse-transcribed using random hexamers, especially in the two GCB-DLBCL cell lines. Conversely, when oligo-dT was used for reverse transcription, GECPAR was clearly detectable in only OCI-LY1, in agreement with the higher abundance of GECPAR in this cell line. (Figure 1D).
GECPAR is predominantly transcribed in germinal center diffuse large B-cell lymphoma cell lines and patients
We measured GECPAR transcription by directional qRT-PCR in 22 DLBCL cell lines (GCB, n=16; ABC, n=8). The overlapping antisense transcript was evaluat- ed in parallel as a control. GECPAR was more frequent- ly expressed in GCB- than ABC-DLBCL cell lines (11/16 vs. 0/8; P=0.001). In particular, it was expressed at high levels in five (OCI-LY1, OCI-LY1b, OCI-LY8, OCI- LY18, VAL), and at lower levels in six (SU-DHL-4, SU- DHL-6, SU-DHL-16, SU-DHL-8, SU-DHL-10, TOLE- DO) GCB-DLBCL cell lines. The transcript was barely detectable in the remaining five GCB and in all the eight ABC-DLBCL cell lines, while the antisense transcript was more broadly expressed in all cell lines (Figure 2A).
We also evaluated GECPAR level in a total RNA-Seq
dataset33 obtained from specimens derived from normal tonsil (n=31) and DLBCL patients (GCB, n=16; ABC, n=18). The transcript was significantly more expressed in normal cells compared to tumor cells, and, in accor- dance with our cell lines data, it was generally more abundant in GCB- than in ABC-DLBCL (Figure 2B). The higher GECPAR expression in GCB-DLBCL was con- firmed in a validation cohort of 74 patients (GCB, n=31; non-GCB, n=43) (GSE145043) (Online Supplementary Figure S2A) and in a second one of 350 patients (GCB, n=183; ABC, n=167) (GSE10846). Variation of GECPAR expression in DLBCL cell lines and patients might be partially explained by its unstable genomic locus.34-36 A focal deletion of the chromosomal region containing the eRNA was observed in three of 737 mature lym- phoid tumors37-41 (Online Supplementary Figure S2B).
The normal tonsil derived cells were then subdivided according to B-cell maturation stage.42 GECPAR was most highly expressed by centroblasts while naïve B cells expressed the lowest levels. This observation fur- ther underlined the specific transcription of GECPAR in GC-derived cells. We also analyzed a catalog of murine lncRNA expressed in different developmental stages of B-cell maturation.43 Similar to our observations in humans, the murine GECPAR orthologue was mainly expressed in GC B cells, confirming the specific and conserved association of GECPAR with the GC B-cell transcriptional program (Online Supplementary Figure S2C).
1134
haematologica | 2022; 107(5)