Page 80 - 2022_02-Haematologica-web
P. 80
O. di Martino et al.
rifice, all but one mouse had developed splenomegaly (aver- age spleen weight 0.5 g) and bone marrow lacked erythroid elements, features that are typical of overwhelming second- ary leukemic engraftment (Online Supplementary Figure S6).
Transcriptional consequences of RXRA DT448/9PP
To assess the maturation effects of RXRA DT448/9PP more comprehensively and to investigate the overlapping
consequences with RXRA WT activation by bexarotene, KMT2A-MLLT3 WT leukemia cells were transduced with RXRA WT or RXRA DT448/9PP and the transcriptional pro- files of the transduced cells were assessed by RNA sequenc- ing (Figure 5A). We noted significant overlap of regulated genes between RXRA WT cells treated with bexarotene and cells transduced with RXRA DT448/9PP (Figure 5B-D). Of 598 genes that were upregulated in RXRA WT cells treated
AB
CD
EF
GH
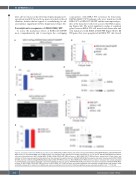
Figure 3. Comparison with other RXRA variants with activity. KMT2A-MLLT3 WT leukemia cells were transduced with MSCV-3xFlag-RXRA R316A/L326A/DT448/9PP- IRES-mCherry or with MSCV-RXRA S427F-IRES-mCherry and evaluated under fluorescent and light microscopy at 72 h (A and E). RXR-KO KMT2A-MLLT3 leukemia cells were transduced with MSCV-3xFlag-RXRA R316A/L326A/DT448/9PP-IRES-mCherry or MSCV-RXRA S427F-IRES-mCherry, stained with FxCycle Violet, and retention of the dye was assessed by flow cytometry at the indicated time points (B and F). RXR-KO KMT2A-MLLT3 leukemia cells were transduced with MSCV-RXRA WT-IRES-mCherry or MSCV-3xFlag-RXRA R316A/L326A/DT448/9PP-IRES-mCherry or MSCV-RXRA S427F-IRES-mCherry, mCherry+ cells were then sorted and plated in methylcellulose, and colonies assessed in technical triplicates on day 7 (C and G). RXR-KO KMT2A-MLLT3 leukemia cells were transduced with MSCV-RXRA-IRES- mCherry or MSCV-RXRA DT448/9PP-IRES-mCherry or MSCV-RXRA S427F-IRES-mCherry and protein expression was evaluated through western blot analysis using anti-RXRA antibody (H-10, Santa Cruz). GAPDH was used as a loading control (D and H). Statistical significance was evaluated using the t-test, ***P<0.001.
422
haematologica | 2022; 107(2)