Page 286 - 2022_01-Haematologica-web
P. 286
A. Astori et al.
1.03, for the 4 donors #3-6), suggesting that RINF knock- down cells could skip one division (in the presence of TGFb). Our data indicate that RINF-dependent erythroid maturation is also dependent on TGFb signaling at physio- logical levels. This cytokine, a potent inducer of erythroid maturation and growth inhibition,27,29,51,52 is known to be enriched in the bone marrow microenvironment53 and we surmise that the RINF/SMAD7 regulation axis that we describe here is likely to exert a more pronounced effect in vivo than in our serum-free culture conditions (i.e., without exogenous TGFb). Moreover, since the mechanism of action is SMAD7-dependent, RINF downregulation may also sensitize hematopoietic cells to other cytokines of the TGFb superfamily which may act on late-stage erythro- poiesis in vivo (such as activins and GDF11).54-57 In this man- ner,RINFcouldbeoneoftheplayersregulatingthedynam-
AB
ic balance between long-term expansion of the erythroid pool versus fast production of RBC for immediate physio- logical needs.
Two recent studies suggest that murine RINF could act as an anchoring platform necessary for TET2 (which lacks a CXXC domain) and its activity at CpG sites (5’- hydroxymethylation), in plasmacytoid dendritic cells15 and mouse embryonic stem cells.48 Conversely, an earlier study indicated that RINF inhibits TET2 and hydrox- ymethylation of genomic DNA in a caspase-dependent manner (in mouse embryonic stem cells), and another one indicated that RINF binds to unmethylated (and not methylated) CpG sites,18 probably reflecting a more com- plex mechanism of action.12 We here provide the first experimental evidence (to our knowledge) that RINF can regulate genome-wide hydroxymethylation in human
C
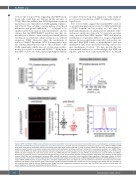
Figure 6. RINF and SMAD7 mRNA closely correlate in primary human CD34+ bone marrow cells isolated from healthy donors or myelodysplastic syndrome patients with del(5q) and RINF-silencing leads to global loss of 5-hydroxymethylation. (A) CD34+ cells were isolated from adult bone marrow (n=11). RINF and SMAD7 mRNA levels were measured by quantitative reverse transcriptase polymerase chain reaction (qRT-PCR). In order to assess whether the expression of these two genes cor- related in donors’ samples, a correlation analysis was performed with the Pearson correlation coefficient method (for non-parametric analysis): rho= 0.684), P(two- tailed) <0.05 (B) RINF and SMAD7 gene expression data were extracted from a previously described microarray dataset published by Pellagatti et al.46 In this study, CD34+ cells were isolated from adult bone marrow (n=17). For mRNA detection, the 233955_x_at and 204790_at probesets were used for CXXC5 and SMAD7, respectively. A correlation analysis was performed with the Pearson rho (for non-parametric analysis). Strong correlation coefficients were noted for normal healthy controls (rho=0.784) and myelodysplastic syndrome (MDS) patients with del(5q), (rho=0.603), which were both highly significant: P(two-tailed) <0.001. (C) CD34+ cells isolated from cord blood or healthy adult donors were transduced with pTripizi vector expressing a shRNA targeting RINF expression (shRNA/RINF#4) or a “non- target” shRNA (shRNA/Control). A couple of days later, during the amplification phase (see Figure 1A), cells were GFP-sorted and 5hmC was detected by immunoflu- orescence (left panel) or flow cytometry (right panel) using an anti-5hmC antibody labeled with a fluorescent dye. The intensity of the labeling was also quantified by cell imaging analysis using Fiji software. Representative images from three independent experiments are shown. Here, the CD34+ cells are from an adult donor at 4 days of expansion (2 days after transduction). Scale bar=50 mm.
278
haematologica | 2022; 107(1)