Page 149 - 2021_09-Haematologica-web
P. 149
Functional and oncogenic roles of PRDM1 in NK cells
AB
CD
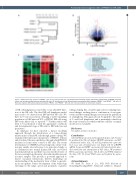
Figure 7. RNA-sequencing analysis of PRDM1-/- cells versus wild-type NK cells. (A) Unsupervised clustering of gene expression profiling data of PRDM1 knock-out clones and cell age-matched parental wild-type NK cells. (B) Volcano plot of the differentiated transcriptional profile between PRDM1-/- and PRDM1+/+ NK cells. (C) Heatmap of genes differentially expressed between PRDM1-/- and PRDM1+/+ NK cells. (D) Summary of gene set enrichment analysis.
of NK-cell malignancies, it probably occurs after EBV infec- tion of the NK cells. The ideal NK-cell lymphoma model may need to be derived from EBV-infected NK cells. We have not been successful in obtaining a viable expanding population of EBV-infected WT or PRDM1 KO cells using EBV from Akata cells as reported.33,34 Further studies will need to be performed to find the appropriate conditions and developmental stage of NK cells for EBV infection and persistence.
In summary, we have reported a disease modeling approach through the introduction of a tumor-driving mutation into normal NK cells through genetic editing. We examined the functional consequences of PRDM1 deletion and elucidated the major pathways through which PRDM1 mediates its homeostatic control of NK cells. The recent development of CRISPR/Cas9 and long-term culture tech- nologies enables selected lesions to be introduced singly or in combination into normal human NK cells. Associated functional alterations can then be assessed in the absence of the noise arising from the many other abnormalities present in tumor samples. This provides a powerful approach to dissect oncogenic interactions, thereby facilitating our understanding of the mechanistic basis of their cooperativ- ity in oncogenesis. Future development of the technology will improve the range, speed and specificity of genetic
editing, making this a feasible approach for studying func- tional changes resulting from a combination of oncogenic events and the essential changes necessary in the generation of a lymphoma. This approach can be applied to the study of T- and B-cell lymphomas and is particularly valuable in the study of tumors for which authentic cell lines or animal models are not available.
Disclosures
No conflicts of interest to disclose.
Contributions
GD and WCC conceived and designed the project; GD, YL and LL performed the NK-cell cloning, CRISPR experiments, and KO cell functional studies; XuL, YS and XiL performed the expression level assay and cell maintenance; CL helped with the CRISPR sgRNA design and HDRT construction; LK helped with the plas- mid construction and identification of KO clones; JW performed RNA-sequencing experiments; AB, JI, QG and TWM analyzed the data; GD, YL, JI and WCC wrote and finalized the manu- script.
Acknowledgments
We thank Dr. Dean A. Lee, MD, PhD (Division of Hematology/Oncology/BMT, Nationwide Children’s Hospital,
haematologica | 2021; 106(9)
2437