Page 205 - 2021_05-Haematologica-web
P. 205
Effects of absence of PTGS1 in platelets
A
B
C
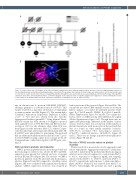
Figure 1. Pedigree phenotype. (A) Pedigree of the affected family, in which black, white and grey symbols indicate presence of the bleeding phenotype, absence of the bleeding phenotype and unknown bleeding phenotype, respectively. The genotype, where known, is shown under each symbol, where G is the mutant allele and C is the reference allele. Double lines indicate consanguinity and strike-through lines are used to indicate deceased individuals. (B) Human Phenotype Ontology (HPO) annotation of the three affected family members. (C) A ribbon diagram of the crystal structure of aspirin-acetylated cyclo-oxygenase 1 (COX-1) showing the location of the variant for the proband which results in a missense substitution of tryptophan to serine at amino acid 322.
ant on chromosome 9, position 125145990 (GRCh37), altering a guanine to a cysteine in exon 8 of PTGS1. This variant resulted in a missense substitution of tryptophan to serine at amino acid 322 (Figure 1C). The variant had a Combined Annotation Dependent Depletion (CADD) score20 of 31.0 and was absent from the Genome Aggregation Database (gnomAD).21 Using Alamut® Visual, the new variant has been shown to be highly conserved with a phyloP score of 9.88. III-3, III-4 and III-5 were het- erozygous (GC) for the alternate allele while IV-2 was homozygous for the reference allele (CC), where C repre- sents the wild-type and G represents the mutant allele. III- 6 and III-7 were unavailable for genotyping. The proband had an additional biallelic mutation (chromosome 7, posi- tion 117175467) causing a splice donor variant in the CF transmembrane conductance regulator (CFTR) gene caus- ing CF.
COX-1 protein in platelets and leukocytes
COX-1 protein in platelet lysates from the proband and her homozygous relatives was absent. In III-3 and III-4, expression was present but reduced and was at normal levels in III-5 and IV-2 (Figure 2A). The absence of COX-1 protein in platelets from the proband and homozygous relatives (Figure 2Ci and Di) compared to a healthy control (Figures 2Bi) was confirmed with immunohistochemical analysis. COX-1 expression, however, was retained in
leukocytes from all those tested (Figure 2Cii and Dii). The variant did not affect COX enzyme activity as shown in kinetic analysis of isolated recombinant protein (wild- type, Km=7.9±0.8 mmol/L; W322S, Km=14.1±1.1 mmol/L, Online Supplementary Figure S1A and B). The variant also had no effect on COX activity after inhibition by aspirin (Online Supplementary Figure S1C). Though there was no appreciable phenotypic difference in the quality of inter- actions observed, there was a reduction in the number of platelet interactions with monocytes in the proband that was not found in other family members (controls, 34.8±19.2%; proband, 7.5%; homozygous relatives, 27.4±9.0%; unaffected relatives, 32.8±10.6%; Figure 3A and C). There was no change in platelet-neutrophil inter- actions (Figure 3B and D).
The role of PTGS1 recessive variant on platelet reactivity
Platelet reactivity is measured in vitro by aggregation and release experiments. Aggregation responses to arachidonic acid (AA; 1 mmol/L) in the proband and her homozygous relatives were reduced compared to control from 65±7% to 4±1%; responses to collagen (1 mg/mL and 3 mg/mL) reduced from 64±13% to 17±10% and from 67±8% to 20±11%, respectively; and responses to adenosine diphos- phate (ADP) at 10 mmol/L was reduced from 62±11% to 38±13%. Interestingly, the proband also had a greatly
haematologica | 2021; 106(5)
1425