Page 133 - 2020_08-Haematologica-web
P. 133
mTORi unlocks AML resistance to LSD1i
LSD1 knockdown significantly increased IRS1 mRNA and protein levels in resistant AML cells, unlike sensitive AML cells (Figure 4C-K and Online Supplementary Figure S8A). Cistrome database analyses of previously deposited LSD1 ChIp-seq tracks12 showed that LSD1 was associated to the IRS1 promoter in resistant NB4 cells, while it was not bound in sensitive KASUMI-1 and SKNO-1 cells (Online Supplementary Figure S8B). We confirmed LSD1 binding to the IRS1 promoter of LSD1i resistant AML cells by ChIP-
qPCR (Figure 4L-M and Online Supplementary Figure S8C- D). Following the induction of IRS1 by DDP38003 treat- ment, H3K4me3, H3K9Ac and H3K27Ac histone marks at IRS1 promoter were strongly increased compared to vehi- cle-treated cells (Figure 4N-P). Modest changes in H3K4me2 were found in THP-1 and KASUMI-1 cells fol- lowing their treatment with LSD1 inhibitors (DDP38003, GSK690 and RN-1) (Online Supplementary Figure S8E-F).
To delineate the hierarchical relationship of IRS1 with
ABC
DEF
GHI
JKL
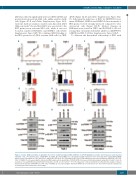
Figure 2. Genetic LSD1 knockdown recapitulates the effects of pharmacological LSD1 inhibition on mTOR signaling in sensitive and irresponsive acute myeloid leukemia cells. (A-C) Growth curves of KASUMI-1 (A), THP-1 (B) and OCI-AML3 (C) cells transduced with retroviral vectors expressing short hairpin RNA (shRNA) against control (scrambled) or LSD1 (shLSD1 #1 and shLSD1 #2) (n=3). (D-F) Normalized LSD1 and CD11b mRNA levels assessed in transduced KASUMI-1 (D), THP- 1 (E), OCI-AML3 (F) cells expressing the indicated shRNA using real-time quantitative PCR (RT-qPCR). Data were statistically analyzed using either Student’s t-test (A, D and G) or one way ANOVA followed by Bonferrroni post hoc test (B, C, E, F, H and I). *: P<0.05 compared to control (scrambled). (J-L) Western blot analysis of lysates obtained from transduced KASUMI-1 (J), THP-1 (K), OCI-AML3 (L) cells expressing the indicated shRNA. The presented blots are derived from replicate samples run on parallel gels and controlled for even loading.
haematologica | 2020; 105(8)
2109