Page 64 - Haematologica - Vol. 105 n. 6 - June 2020
P. 64
L.A. Richards et al.
the DKC1 promoter.26,27 ChIP sequencing data from peripheral blood erythroblasts made available through ENCODE on the UCSC browser also provide evidence of GATA 1 binding at the DKC1 promoter, in the vicinity of -1097 to -493 relative to the transcription start site (chrX: 153,991,030, hg19) (Figure 5B). Guided by these data, we identified putative GATA sites at positions: -679 to -668 and -453 to -468 and designed PCR primers to interrogate the transcription factor binding by ChIP. First, using antibodies to trimethylated H3K4 (H3K4me3), which occupies transcriptionally active chromatin, and H3K27me3, which identifies repressed sites, we verified that chromatin at the DKC1 promoter was in an open configuration conducive to transcriptional activation in both STF-stimulated cells and erythroblastic cells (Figure 5C).29 ChIP analysis also confirmed MYC binding to the DKC1 promoter, although this appeared to diminish pro- gressively as undifferentiated CB cells underwent ery- throid differentiation in SE culture. Conversely, GATA1 binding at the DKC1 promoter appeared most robust at week 3, corresponding with the accumulation of ery-
throblasts expressing high levels of GLYA (Online Supplementary Figure S5) and peak expression of DKC1 in six out of nine CB cultures (Online Supplementary Figure S2). Consistent with GATA1 binding in GLYAhigh CB- derived erythroblasts (Figure 5C), ChIP sequencing results from three independent investigations, accessed using CistromeDB, showed GATA1 binding at the DKC1 promoter of erythroblasts derived from bone marrow and peripheral blood (Online Supplementary Figure S6).30-32 Although TAL1 can bind DNA via E-boxes, no TAL1 binding at the DKC1 promoter was detected in CB cells at any stage of culture. Collectively, these data suggest a model whereby MYC binds the DKC1 promoter in undifferentiated cells and is replaced by GATA1 during erythroid differentiation.
Since GATA1 regulation of DKC1 has not previously been described, a luciferase reporter assay was conducted to confirm that GATA sites contribute to DKC1 transcrip- tion. Mutations were induced in two potential GATA1 binding sites of the DKC1 promoter construct by site- directed mutagenesis and luciferase activity was measured
ABE
CD
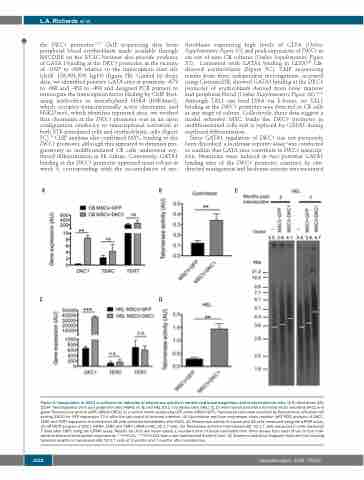
Figure 3. Upregulation of DKC1 is sufficient for induction of telomerase activity in normal cord blood progenitors and erythroleukemia cells. (A-D) Cord blood (CB) CD34+ hematopoietic stem and progenitor cells (HSPC) (A, B) and HEL 92.1.7 leukemia cells (HEL) (C, D) were transduced with a lentiviral vector encoding DKC1 and green fluorescence protein (GFP) (MSCV-DKC1) or a control vector expressing GFP alone (MSCV-GFP). Transduced cells were enriched by fluorescence activated cell sorting (FACS) for GFP expression 72 h after the last round of lentiviral infection. (A) Quantitative real-time polymerase chain reaction (qRT-PCR) analysis of DKC1, TERC and TERT expression in transduced CB cells collected immediately after FACS. (B) Telomerase activity in transduced CB cells measured using the qTRAP assay. (C) qRT-PCR analysis of DKC1 mRNA, TERC and TERT mRNA in HEL 92.1.7 cells. (D) Telomerase activity in transduced HEL 92.1.7 cells measured in cells harvested 7 days after FACS using the qTRAP assay. Results for (A-D) are mean values ± standard error of mean calculated from three assays from each of two to four inde- pendent lentiviral transduction experiments. **P<0.01, ***P<0.001 from a two-tailed paired Student t test. (E) Telomeric restriction fragment Southern blot showing telomere lengths in transduced HEL 92.1.7 cells at 2 months and 7 months after transduction.
1522
haematologica | 2020; 105(6)