Page 40 - Haematologica - Vol. 105 n. 6 - June 2020
P. 40
G. Pavlasova and M. Mraz
induced in CLL cells treated by microenvironmental fac- tors such as IL4, TNFα, INFα or GMCSF in vitro45-47(and our unpublished data).
CD20 silencing in malignant B cells revealed that CD20 affects the phosphorylation of multiple BCR-associated kinases and proteins after BCR-ligation (LYN, SYK, GAB1, and ERK).34 This suggests that both CD20 and BCR are induced in immune niches34 to allow effective and strong BCR activation by an antigen or CD20 might also be involved in some form of “tonic” BCR signaling.48 This has important implications for combining BCR inhibitors with antibodies targeting CD20. We have shown that inhibit- ing BTK interferes with CXCR4 signaling in CLL cells and thus leads to very significant repression of CD20 expres- sion in CLL cells. This might partially explain the lack of clinical benefit from adding rituximab to ibrutinib.12 Ibrutinib was recently tested and approved in combina- tion with a more potent anti-CD20 monoclonal antibody, namely obinutuzumab,49 whose efficacy is less affected by lower levels of CD20 on the cell-surface. We also suggest that PI3K inhibition, like BTK inhibition, might lead to the downmodulation of CD20, but this remains to be formal- ly proven, and the implications for the therapeutic combi- nation of rituximab with PI3K inhibitors (including idelal- isib) or other BTK inhibitors (such as acalabrutinib) are unclear.
Altogether, functional studies suggest that CD20 is physiologically directly required for efficient BCR signal- ing in B cells. This is also in line with some data from the CD20 mouse models. In Uchida’s CD20-/- mice model, cell-surface IgM expression on both mature and immature B cells was 20 – 30% lower than that on B cells from wild- type littermates, which was connected with reduced BCR- and CD19-dependent intracellular calcium mobilization.36 In a study by Morsy et al., the reduction in BCR-associated calcium mobilization in CD20-/- murine B cells was pro- posed to be caused by a defect in calcium transport rather than in its release from intracellular stores.38 In our opin- ion, there is a sufficient body of evidence suggesting that CD20 is involved in BCR signaling, but it is unclear whether this is related to its putative function as a calcium channel and/or other function(s). Similarly, it is not clear if other molecular pathways or B-T-cell interactions might be affected by CD20 levels on the cell-surface of B cells.
Regulation of CD20 transcription and its "therapeutic modulation"
Rituximab is one of the most effective and widely used therapeutic monoclonal antibodies, but malignant B cells can become relatively resistant to such therapy. Mechanisms of malignant B cells’ resistance to anti-CD20 monoclonal antibodies include insufficient CDC activity due to increased expression of regulatory proteins CD55, CD59 or factor H,50,51 less effective ADCC in cases with specific FcγRIII polymorphism,52 exhaustion of cytotoxic mechanisms (such as complement/effector cells),53,54 poly- morphism in the complement component C1qA,55 or abnormal composition and localization of lipid rafts and thus impaired rituximab-induced apoptosis (Figure 1).56 Nevertheless, one of the most straightforward and fre- quent causes of resistance to anti-CD20 monoclonal anti- bodies is reduced CD20 expression, which can be due to (de)regulation of transcriptional, post-transcriptional, or post-translational mechanisms (including CD20 protein transport to the cell surface57).
Regarding transcriptional regulation, the MS4A1 gene lacks several regulatory elements typical of other B-cell specific genes, including TATA and CAAT box. The
Table 1. Positive and negative regulators of MS4A1 (CD20 gene) tran- scription.
Positive regulatorsRef
Transcriptionfactors USF, TFE359
OCT1,58 OCT258 PU.1/PiP (IRF4)59
ELK1,64 ETS164 SP165
NFκB65
CHD4,69* MBD269*
Epigenetic regulators Unknown
Negative regulatorsRef
FOXO170 CREM69
SMAD2/399 MYC67
Sin3A-HDAC176 EZH280 HDAC1/481 HDAC682
RBPJ and mutated NICD of NOTCH195
* The DNA binding site in the MS4A1 promoter has not been defined. NICD: NOTCH1 intracellular domain.
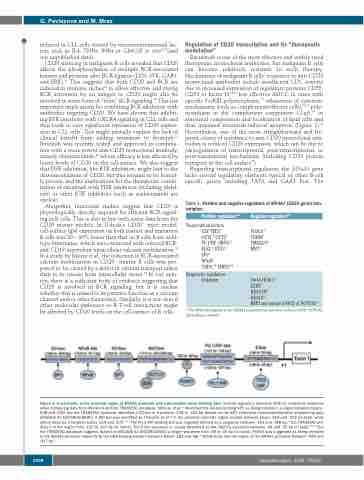
Figure 3. A schematic of the proximal region of MS4A1 promoter with transcription factor binding sites. Several regulatory elements differ in nucleotide sequence when comparing data from literature and the TRANSFAC database. Shimizu et al.65 described the GC-box binding SP1 as being located in a region between bases - 548 and -539, but the TRANSFAC database identified a GC-box in a position -234 to -224 bp [based on the SP1 chromatin immunoprecipitation sequencing data (ENCODE ID: ENCSR000BHK)]. A BAT-box was identified by Thévenin et al.58 in the proximal promoter region located between bases -214 and -202 (in bold), while others describe it between bases -214 and -200.59,65 The PU.1/PiP binding site was originally defined as a sequence between -161 and -148 bp,59 but TRANSFAC pre- dicts it in the region from -120 to -107 bp (in italics). The E-box sequence is usually described as the CACCTG sequence between -44 and -39 bp (in bold)59,62,65 but the TRANSFAC database suggests (based on ENCODE ID: ENCSR000BGI) a longer sequence from -48 to -35 bp (in italics). FOXO1 was suggested as being recruited to the MS4A1 promoter indirectly by the DNA-binding element between bases -182 and -88.70 NFκB binds into the region of the MS4A1 promoter between -425 and -417 bp.62
1498
haematologica | 2020; 105(6)