Page 156 - Haematologica - Vol. 105 n. 6 - June 2020
P. 156
J.A. Cohen et al.
Introduction
Clinical staging using the Binet and Rai classification systems provides a simple and inexpensive approach to assess prognosis in chronic lymphocytic leukemia (CLL).1,2 However, most patients today are diagnosed in early stages of the disease (Binet A or Rai 0) when these prog- nosticators fail to provide adequate risk stratification.3 Although similarly classified as early-stage CLL, Binet A and Rai 0 patients demonstrate heterogeneous clinical courses ranging from normal life expectancy in the absence of any treatment to unexpectedly short progres- sion-free intervals rapidly requiring clinical intervention.4
To overcome the inherent weakness of clinical staging systems, other parameters have been sought and pro- posed by several studies as reliable prognosticators in CLL, including immunocytogenetic and molecular mark- ers such as deletions of the short arm of chromosome 17 (del17p) and mutations of the TP53 gene, deletions of the long arm of chromosome 11 (del11q), and trisomy of chro- mosome 12 (tris12), the immunoglobulin heavy chain (IGHV) gene mutational status as well as biochemical parameters such as beta-2-microglobulin (B2M) and thymidine kinase (TK) and cell surface receptors such as the integrin CD49d.5-10
Novel prognostic indices and model systems have been
developed to integrate these markers into comprehensive
scoring systems, such as the CLL International Prognostic
Index (CLL-IPI), the German CLL Study Group (GCLLSG)
index, and the MD Anderson Cancer Center (MDACC)
score.11-14 Although validations in the setting of early-stage
CLL and/or treatment-free-survival (TFS) prediction have
been undertaken, these indices were originally generated
to predict overall survival operating across all stages of dis- ease.11,13,15,16
Here we present a novel laboratory-based prognostic index specifically developed to predict TFS in Rai 0 CLL, thus allowing clinicians and researchers to uniformly and more accurately identify cases with higher risk for need- ing early treatment.
Methods
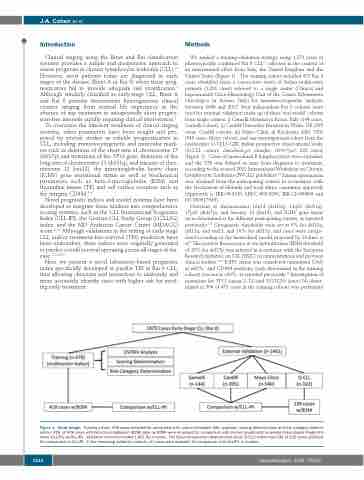
We applied a training-validation strategy using 1,879 cases of phenotypically confirmed Rai 0 CLL17 collected in the context of an international effort from Italy, the United Kingdom and the United States (Figure 1). The training cohort included 478 Rai 0 cases identified from a consecutive series of Italian multicenter patients (1,201 cases) referred to a single center (Clinical and Experimental Onco-Hematology Unit of the Centro Riferimento Oncologico in Aviano, Italy) for immunocytogenetic analyses between 2006 and 2017. Four independent Rai 0 cohorts were used for external validation made up of three ‘real world’ cohorts from single centers, i) Gemelli Hospital in Rome, Italy (144 cases, Gemelli cohort), ii) Cardiff University Hospital in Wales, UK (395 cases, Cardiff cohort), iii) Mayo Clinic in Rochester, MN, USA (540 cases, Mayo cohort), and one investigational cohort from the multicenter O-CLL1-GISL Italian prospective observational study (O-CLL cohort; clinicaltrial.gov identifier: 00917540; 322 cases) (Figure 1). Cases of monoclonal B lymphocytosis were excluded, and the TFS was defined as time from diagnosis to treatment, according to the revised 2018 International Workshop on Chronic Lymphocytic Leukemia (IWCLL) guidelines.18 Patient information was obtained from the participating centers in accordance with the Declaration of Helsinki and local ethics committee approvals (Approvals n. IRB-05-2010, LREC #02/4806, IRB-12-000969 and NCT00917540).
Deletions at chromosomes 13q14 (del13q), 11q23 (del11q), 17p13 (del17p), and trisomy 12 (tris12), and IGHV gene status were determined at the different participating centers, as reported previously.5,19 Cytogenetic thresholds were set at 5% for del13q, del11q, and tris12, and 10% for del17p, and cases were catego- rized according to the hierarchical model proposed by Dohner et al.7 The positive fluorescence in situ hybridization (FISH) threshold of 10% for del17p was selected in accordance with the European Research Initiative on CLL (ERIC) recommendations and previous clinical studies.20-22 IGHV status was considered unmutated (UM) at ≥98%,6 and CD49d positivity (only determined in the training cohort) was set at >30%, as reported previously.19 Investigation of mutations for TP53 (exons 2–11) and NOTCH1 (exon 34) (deter- mined in 304 of 478 cases in the training cohort) was performed
Figure 1. Study design. Training cohort: 478 cases included for univariable (UV) and multivariable (MV) analyses, scoring determination and risk category determi- nation; 418 of 478 cases with beta-2-microglobulin (B2M) data (w/B2M) were employed for comparison with chronic lymphocytic leukemia International Prognostic Index (CLL-IPI) (w/CLL-IPI). Validation cohorts included 1,401 Rai 0 cases. The Italian prospective observational study (O-CLL) cohort had 239 of 322 cases available for comparison to CLL-IPI. In the remaining validation cohorts, all cases were available for comparison with CLL-IPI. n: number.
1614
haematologica | 2020; 105(6)