Page 152 - Haematologica - Vol. 105 n. 6 - June 2020
P. 152
S. Ferrero et al.
AB
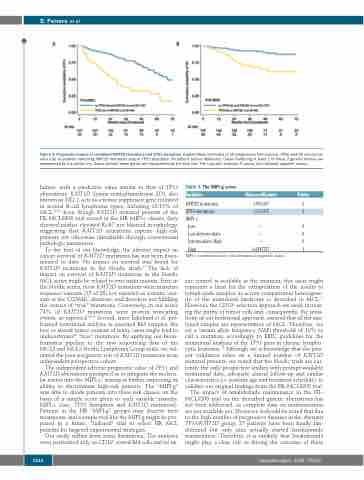
Figure 4. Prognostic impact of combined KMT2D mutations and TP53 disruption. Kaplan-Meier estimates of (A) progression free survival (PFS) and (B) overall sur- vival (OS) of patients harboring KMT2D mutations and/or TP53 disruption (mutations and/or deletions). Cases harboring at least 1 of these 3 genetic lesions are represented by the yellow line. Cases without these genes are represented by the blue line. The Log-rank statistics P-values are indicated adjacent curves.
failure, with a predictive value similar to that of TP53 aberrations. KMT2D (lysine methyltransferase 2D), also known as MLL2, acts as a tumor suppressor gene mutated in several B-cell lymphoma types, including 10-15% of MCL.28-31 Even though KMT2D mutated patients of the FIL-MCL0208 trial scored in the HR MIPI-c classes, they showed neither elevated Ki-67 nor blastoid morphology, suggesting that KMT2D mutations capture high-risk patients not otherwise identifiable through conventional pathologic parameters.
To the best of our knowledge, the adverse impact on cancer survival of KMT2D mutations has not been docu- mented to date. No impact on survival was found for KMT2D mutations in the Nordic study.17 The lack of impact on survival of KMT2D mutations in the Nordic MCL series might be related to two main reasons. First, in the Nordic series, most KMT2D mutations were missense sequence variants (15 of 28) not reported as somatic vari- ants in the COSMIC database, and therefore not fulfilling the criteria of “true” mutations. Conversely, in our series 74% of KMT2D mutations were protein truncating events, as expected.28-31 Second, since Eskelund et al. per- formed mutational analysis in unsorted BM samples, the low or absent tumor content of many cases might lead to underestimate28 “true” mutations. By applying our bioin- formatics pipeline to the raw sequencing data of the MCL2 and MCL3 Nordic Lymphoma Group trials, we val- idated the poor prognostic role of KMT2D mutations in an independent prospective cohort.
The independent adverse prognostic value of TP53 and KMT2D aberrations prompted us to integrate the molecu- lar results into the MIPI-c,8 aiming at further improving its ability to discriminate high-risk patients. The “MIPI-g” was able to divide patients into three risk classes, on the basis of a simple score given to each variable (namely: MIPI-c class, TP53 disruption and KMT2D mutations). Patients in the HR “MIPI-g” groups may deserve new treatments, and a simple tool like the MIPI-g might be pro- posed in a future, “tailored” trial to select HR MCL patients for targeted experimental strategies.
Our study suffers from some limitations. The analyses were performed only on CD19+ sorted BM cells and no tis-
Table 3. The MIPI-g score. Variables
KMT2D mutations TP53 disruption MIPI-c
Beta-coefficients
Points
1,113,875 2 Low − 0
1,035,607 2
Low-Intermediate
Intermediate-High
High
− 0
− 0 0.6847757 1
MIPI-c: combined mantle cell international prognostic index.
sue control is available at the moment; this issue might represent a limit for the extrapolation of the results to lymph-node samples, as across-compartment heterogene- ity of the mutational landscape is described in MCL.10 However, the CD19+ selection approach we used, increas- ing the purity of tumor cells and, consequently, the sensi- tivity of our mutational approach, ensured that all the ana- lyzed samples are representative of MCL. Therefore, we set a variant allele frequency (VAF) threshold of 10% to call a mutation, accordingly to ERIC guidelines for the mutational analysis of the TP53 gene in chronic lympho- cytic leukemia.32 Although we acknowledge that the pres- ent validation relies on a limited number of KMT2D mutated patients, we noted that the Nordic trials are cur- rently the only prospective studies with prompt available mutational data, adequate clinical follow-up and similar characteristics (i.e. patients age and treatment schedule), to validate our original findings from the FIL-MCL0208 trial.
The impact of lenalidomide maintenance in the FIL- MCL0208 trial on the described genetic aberrations has not been addressed, as complete data on randomization are not available yet. However, it should be noted that due to the high number of progressive diseases in the aberrant TP53/KMT2D group, 27 patients have been finally ran- domized but only nine actually started lenalidomide maintenance. Therefore, it is unlikely that lenalidomide might play a clear role in driving the outcome of these
1610
haematologica | 2020; 105(6)