Page 131 - Haematologica - Vol. 105 n. 6 - June 2020
P. 131
PTCL classification using RT-MLPA assay
Discussion
The classification of PTCL is often challenging and poor- ly reproducible, with a recent study showing a 31.5% rate of discrepancy between the referral and expert diagnoses,4 likely due to the complexity of these rare neoplasms and the wide range of practices among pathologists and labora- tories.3 Hsi et al. pointed out the limited number of immunohistochemical markers assessed in routine prac-
A
tice, especially the TFH markers, resulting in a poor charac- terization of PTCL and a high frequency of PTCL-NOS diagnosed in the US.22 The ligation-dependent RT-PCR assay has been reported to be a simple and robust assay applicable to FFPE samples that can be used to classify DLBCL into GCB or ABC subtypes.23,24 Here, we expanded on this as RT-MLPA can contribute to classify the main specified categories of non-cutaneous PTCL in routine practice. This assay, which can be performed relatively
B
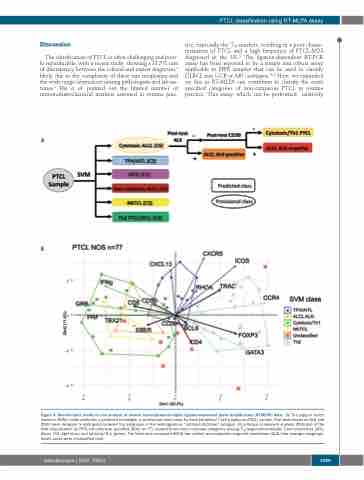
Figure 4. Bioinformatic model for the analysis of reverse transcriptase-multiplex ligation-dependent probe amplification (RT-MLPA) data. (A) The support vector machine (SVM) model attributes a predicted (rectangle) or provisional (oval) class for each peripheral T-cell lymphoma (PTCL) sample. Post-tests based on ALK and CD30 were designed to distinguish between the subgroups in the heterogeneous “cytotoxic/ALCL-like” category. (B) principal component analysis (PCA) plot of the SVM classification for PTCL-not otherwise specified (NOS) (n=77) showed three main molecular categories among: TFH/angioimmunoblastic T-cell lymphomas (AITL) (blue), Th2 (light blue), and cytotoxic/Th1 (green). The latter also comprised NKTCL-like (yellow) and anaplastic large cell lymphomas (ALCL)-like (orange) subgroups. Seven cases were unclassified (red).
haematologica | 2020; 105(6)
1589