Page 119 - Haematologica - Vol. 105 n. 6 - June 2020
P. 119
DNA methylation in T-ALL
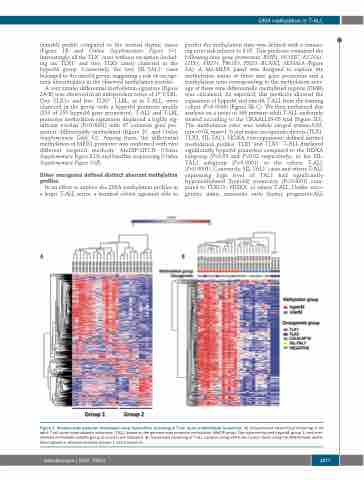
(interM) profile compared to the normal thymic tissue (Figure 1B and Online Supplementary Figure S1). Interestingly, all the TLX+ cases without exception (includ- ing six TLX1+ and two TLX3+ cases) clustered in the hyperM group. Conversely, the two SIL-TAL1+ cases belonged to the interM group; suggesting a role of oncoge- netic abnormalities in the observed methylation profiles.
A very similar differential methylation signature (Figure 2A-B) was observed in an independent series of 17 T-LBL. One TLX1+ and five TLX3+ T-LBL, as in T-ALL, were clustered in the group with a hyperM promoter profile (253 of 255 hyperM gene promoters). T-ALL and T-LBL promoter methylation signatures displayed a highly sig- nificant overlap (P<0.0001) with 97 common gene pro- moters differentially methylated (Figure 2C and Online Supplementary Table S2). Among them, the differential methylation of MEIS1 promoter was confirmed with two different targeted methods, MeDIP-QPCR (Online Supplementary Figure S2A) and bisulfite sequencing (Online Supplementary Figure S2B).
Driver oncogenes defined distinct aberrant methylation profiles
In an effort to explore the DNA methylation profiles in a larger T-ALL series, a minimal robust signature able to
AB
predict the methylation state was defined with a remain- ing error risk inferior to 0.05. This predictor contained the following nine gene promoters: BMP4, HOXB7, KCNA1, LHX1, MEIS1, PROX1, PSD3, RUNX2, SEMA6A (Figure 3A). A MS-MLPA panel was designed to explore the methylation status of these nine gene promoters and a methylation ratio corresponding to the methylation aver- age of these nine differentially methylated regions (DMR) was calculated. As expected, this predictor allowed the separation of hyperM and interM T-ALL from the training cohort (P=0.0016) (Figure 3B-C). We then performed this analysis on a series of 168 primary adult T-ALL uniformly treated according to the GRAALL03-05 trial (Figure 3D). The methylation ratio was widely ranged (mean=0.62, min=0.04, max=1.1) and major oncogenetic drivers (TLX1, TLX3, SIL-TAL1, HOXA overexpression) defined distinct methylation profiles. TLX1+ and TLX3+ T-ALL displayed significantly hyperM promoters compared to the HOXA subgroup (P=0.03 and P=0.02 respectively), to the SIL- TAL1 subgroup (P<0.0001) or the others T-ALL (P<0.0001). Conversely, SIL-TAL1+ cases and others T-ALL expressing high level of TAL1 had significantly hypomethylated (hypoM) promoters (P<0.0001) com- pared to TLX1/3+, HOXA+ or others T-ALL. Unlike onco- genetic status, immature early thymic progenitor-ALL
Figure 1. Genome-wide promoter methylation-array hierarchical clustering in T-cell acute lymphoblastic leukemias. (A) Unsupervised hierarchical clustering of 24 adult T-cell acute lymphoblastic leukemias (T-ALL) based on the genome-wide promoter methylation (MeDIP-array). The hypermethylated (hyperM; group 1) and inter- mediate methylated (interM; group 2) clusters are indicated. (B) Supervised clustering of T-ALL samples along with three human thymi using the differentially methy- lated signature obtained between groups 1 and 2 (panel A).
haematologica | 2020; 105(6)
1577