Page 107 - Haematologica May 2020
P. 107
BCR-FGFR1 regulated by dimerization and chaperonin Hsp90
A
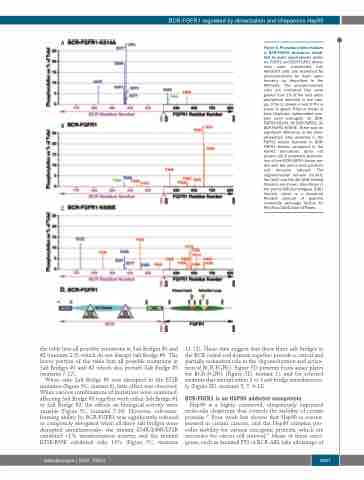
Figure 4. Phosphorylated residues in BCR-FGFR1 derivatives identi- fied by mass spectrometry analy- sis. FGFR1 and BCR-FGFR1 deriva-
B
C
tives were
HEK293T cells and examined for phosphorylation by mass spec- trometry as described in the Methods. The phosphorylation sites are indicated that were greater than 1% of the total phos- phorylation detected in the sam- ple. P-Tyr is shown in red; P-Thr is shown in green; P-Ser is shown in blue. Duplicate, independent sam- ples were averaged; (A) BCR- FGFR1-K514A, (B) BCR-FGFR1, (C) BCR-FGFR1-K656E. There was no significant difference in the phos- phorylation sites detected in the FGFR1 kinase domains in BCR- FGFR1 fusions compared to the FGFR1 derivatives (data not shown). (D) A schematic presenta- tion of the BCR-FGFR1 fusion pro- tein with key amino acid positions and domains labeled. The oligomerization domain (OLIGO), the Grb2 and the Abl SH2 binding domains are shown. Also shown is the partial DBL-homologous (DBL) domain, which is a structural RhoGEF domain of guanine nucleotide exchange factors for Rho/Rac/Cdc42-like GTPases.
transfected into
the table lists all possible mutations in Salt Bridges #1 and #2 (mutants 2-5) which do not disrupt Salt Bridge #3. The lower portion of the table lists all possible mutations in Salt Bridges #1 and #2 which also perturb Salt Bridge #3 (mutants 7-12).
When only Salt Bridge #3 was disrupted in the E52R mutation (Figure 5C, mutant 6), little effect was observed. When various combinations of mutations were examined, affecting Salt Bridge #3 together with either Salt Bridge #1 or Salt Bridge #2, the effects on biological activity were variable (Figure 5C, mutants 7-10). However, cell-trans- forming ability by BCR-FGFR1 was significantly reduced or completely abrogated when all three salt bridges were disrupted simultaneously: the mutant E34R/E46R/E52R exhibited <1% transformation activity, and the mutant R53E/R55E exhibited only 14% (Figure 5C, mutants
11-12). These data suggest that these three salt bridges in the BCR coiled-coil domain together provide a critical and partially redundant role in the oligomerization and activa- tion of BCR-FGFR1. Figure 5D presents focus assay plates for BCR-FGFR1 (Figure 5D, mutant 1), and for selected mutants that disrupt either 2 or 3 salt bridge simultaneous- ly (Figure 5D, mutants 5, 7, 9-12).
BCR-FGFR1 is an HSP90 addicted oncoprotein
Hsp90 is a highly conserved, ubiquitously expressed molecular chaperone that controls the stability of certain proteins.26 Prior work has shown that Hsp90 is overex- pressed in certain cancers, and the Hsp90 complex pro- vides stability for various oncogenic proteins, which are necessary for cancer cell survival.27 Many of these onco- genes, such as mutated P53 or BCR-ABL take advantage of
haematologica | 2020; 105(5)
1267