Page 40 - Haematologica April 2020
P. 40
D. Wu et al.
splicing, leading to protein dysfunction (see Online Supplementary Table S3 of reference by Luo and Feurstein18). All of the three canonical splicing site vari- ants in the pilot set were classified as PATH or LPATH. More challenging, however, is the consideration of syn- onymous/intronic/non-coding variants which may result in cryptic splice site activation, and/or enhancement or repression of adjacent canonical splice sites. For example, the intronic NM_001754:c.508+3delA variant has been reported in a single family with disease segregation (8 meioses, PP1_strong). Several family members were diag- nosed with thrombocytopenia, aspirin-like platelet aggre- gation defects, and dense granule abnormalities.49 This variant is absent from population databases (PM2) and both splicing predictors (MaxEntScan and Splice- SiteFinder)50,51 predict a significant decrease in the score of the canonical splice site (PP3). Moreover, experimental reverse transcriptase polymerase chain reaction studies (PS3), using RNA derived from two affected family mem- bers, were performed and indicate the creation of a novel cryptic splice site 23 nucleotides upstream of the normal splice site resulting in a frameshift p.(Arg162fs*177), and the transcript is predicted to undergo nonsense-mediated decay.49 Combining all of these codes, a final assertion of PATH is given by the MM-VCEP (Table 3).
BP7 is a benign code specifically designed to evaluate syn- onymous/intronic/non-coding variants in the ACMG/AMP framework. BP7 can be applied if computational evidence suggests no impact on splicing, and the nucleotide is not conserved. The ClinVar variant with conflicting interpreta- tions in ClinVar, NM_001754:c.444C>T, p.(Thr148=), has been classified as LBEN by the MM-VCEP using BP7 and the benign in silico prediction code, BP4 (Table 3). This nucleotide change is predicted to have no impact on splic- ing and it is also not conserved (phyloPscore: -4.3832, below the MM-VCEP-specified threshold of <0.118). Clinical data from seven individuals with this variant were acquired from the original ClinVar submitter (SCV000761123.1) and revealed that none of the probands met any of the RUNX1 phenotypic criteria.18 Currently, only two RUNX1 variants have been reported to display an abnormal splicing effect as demonstrated by RNA assays.11,49 The potential effects of other splicing variants rely solely on in silico predictions. Although there is robust effort in consideration of algorithms to predict the effects of splicing variants, these algorithms require further evaluation. Indeed, we know of only limited experimental data within the RUNX1 gene specifically to test these tools. Accordingly, the synonymous variant, NM_001754:c.1257G>A, p.(Val419=) is predicted to cre- ate alternative splice acceptor sites, but is not expected to abolish any existing consensus sites, as it is too far away from either end of the exon. Due to this in silico prediction result, none of the PP3/BP4 and BP7 codes can be assigned, and the classification of this variant remains a VUS. Further resolution of the significance of this variant could be obtained through parental testing, and/or RNA- sequencing data.
Example 5. Copy number variants, deletion of exon 2 (PATH with PVS1_moderate, PM2, PS4, and PP1_strong)
Not infrequently, patients with FPD/AML have been reported to have copy number variants resulting in intra- genic deletions of RUNX1.52 As part of our pilot cohort, we evaluated several probands with copy number vari-
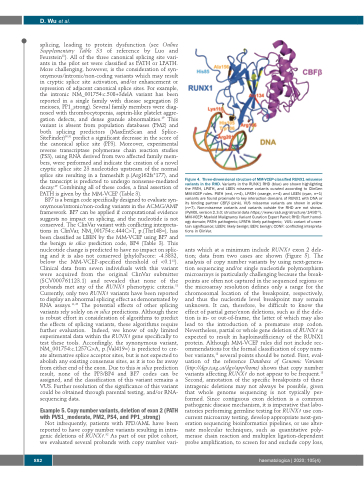
Figure 4. Three-dimensional structure of MM-VCEP-classified RUNX1 missense variants in the RHD. Variants in the RUNX1 RHD (blue) are shown highlighting the PATH, LPATH, and LBEN missense variants curated according to ClinGen MM-VCEP rules. PATH (red, n=4), LPATH (orange, n=4) and LBEN (cyan, n=1) variants are found proximate to key interaction domains of RUNX1 with DNA or its binding partner CBFβ (pink). VUS missense variants are shown in yellow (n=7). Non-missense variants and variants outside the RHD are not shown. (PyMOL version 2.3.0; structural data https://www.rcsb.org/structure/1H9D 82). MM-VCEP: Myeloid Malignancy Variant Curation Expert Panel; RHD: Runt homol- ogy domain; PATH: pathogenic; LPATH: likely pathogenic; VUS: variant of uncer- tain significance; LBEN: likely benign; BEN: benign; CONF: conflicting interpreta- tions in ClinVar.
ants which at a minimum include RUNX1 exon 2 dele- tion; data from two cases are shown (Figure 5). The analysis of copy number variants by using next-genera- tion sequencing and/or single nucleotide polymorphism microarrays is particularly challenging because the break- points are often not captured in the sequenced regions or the microarray resolution defines only a range for the chromosomal location of the breakpoint, respectively, and thus the nucleotide level breakpoint may remain unknown. It can, therefore, be difficult to know the effect of partial gene/exon deletions, such as if the dele- tion is in- or out-of-frame, the latter of which may also lead to the introduction of a premature stop codon. Nevertheless, partial or whole gene deletion of RUNX1 is expected to result in haploinsufficiency of the RUNX1 protein. Although MM-VCEP rules did not include rec- ommendations for the formal classification of copy num- ber variants,18 several points should be noted. First, eval- uation of the reference Database of Genomic Variants (http://dgv.tcag.ca/dgv/app/home) shows that copy number variants affecting RUNX1 do not appear to be frequent.18 Second, annotation of the specific breakpoints of these intragenic deletions may not always be possible, given that whole genome sequencing is not typically per- formed. Since contiguous exon deletion is a common pathogenic disease mechanism, it is imperative that labo- ratories performing germline testing for RUNX1 use con- current microarray testing, develop appropriate next-gen- eration sequencing bioinformatics pipelines, or use alter- nate molecular techniques, such as quantitative poly- merase chain reaction and multiplex ligation-dependent probe amplification, to screen for and exclude copy loss,
882
haematologica | 2020; 105(4)