Page 32 - Haematologica April 2020
P. 32
D. Wu et al.
874
haematologica | 2020; 105(4)
continued from the previous page
ACMG/ AMP CC
Original ACMG/AMP rule summary
Specification
Stand alone
Very strong
Strong
Moderate
Supporting
Comments
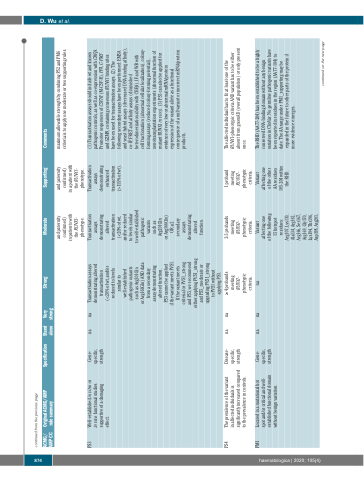
PS3
Well-established in vitro or in vivo functional studies supportive of a damaging effect.
Gene- specific, strength
na
na
Transactivation assays demonstrating altered transactivation (<20% of wt, and/or reduced to levels similar to well-established pathogenic variants such as Arg201Gln
or Arg166Gln) AND data from a secondary assay demonstrating altered function.
PS3 cannot be applied
if the variant meets PVS1. If the variant meets criteria for PVS1_strong and PS3, we recommend either applying PVS1_strong and PS3_moderate or upgrading PVS1_strong to PVS1 without applying PS3.
Transactivation assays demonstrating altered transactivation (<20% of wt, and/or reduced to levels similar
Transactivation assays demonstrating enhanced transactivation (>115% of wt).
(1) Transactivation assays should include wt and known pathogenic controls, as well as co-expression with CBFβ. Promoter sequences of CSF1R (M-CSF-R), PF4, C-FMS and GZMB, containing consensus RUNX1 binding sites have been used for transactivation assays. (2) The following secondary assays have been performed: EMSA and yeast hybrid assays (decreased DNA-binding affinity), co-IP, FRET and affinity assays (diminished heterodimerization ability with CBFβ), IF and WB with cell fractionation (abnormal cellular localization), colony- forming assays (reduced colony-forming potential), xenotransplantation experiments (abnormal function of mutant RUNX1 in vivo). (3) PS3 can also be applied for evidence of very low or abnormal mRNA/protein expression of the variant allele as a functional consequence of a null variant or incorrect mRNA/protein products.
PS4
The prevalence of the variant
in affected individuals is significantly increased compared to the prevalence in controls.
Disease- specific, strength
na
na
≥ 4 probands meeting RUNX1- phenotypic criteria.
2-3 probands meeting RUNX1- phenotypic criteria.
1 proband meeting RUNX1- phenotypic
The affected individual has to fit at least one of the RUNX1-phenotypic criteria AND variant has to be either absent from gnomAD (overall population) or only present once.
PM1
Located in a mutational hot spot and/or critical and well- established functional domain without benign variation.
Gene- specific, strength
na
na
na
Variant affecting one of the following 13 hotspot residues: Arg107, Lys110, Ala134, Arg162, Arg166, Ser167, Arg169, Gly170, Lys194, Thr196, Asp198, Arg201,
Variant affecting one of the other AA residues 105-204 within the RHD.
The RHD (AA 77-204) has been established to be a highly conserved DNA-binding domain without any benign variation in ClinVar. No germline pathogenic variants have been reported in residues in the region (AA 77-104) to date. The AA range under PM1_supporting may be
and paternity confirmed) in patients with the RUNX1- phenotype.
and paternity confirmed) in a patient with the RUNX1- phenotype.
maximum allowable strength by combining PS2 and PM6 criteria is to apply one moderate or two supporting rules.
to well-established pathogenic variants
such as Arg201Gln
or Arg166Gln) OR ≥2 secondary assays demonstrating altered function.
criteria.
expanded in the future to other parts of the protein if more evidence emerges.
continued on the next page