Page 31 - Haematologica April 2020
P. 31
RUNX1 variant curation
haematologica | 2020; 105(4)
873
continued from the previous page
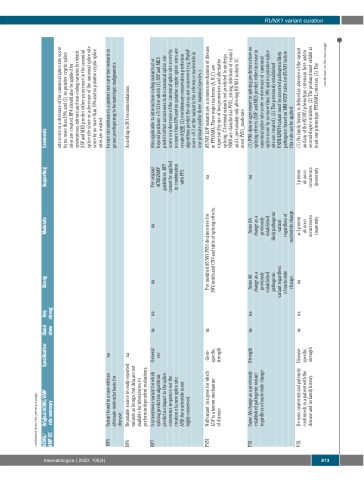
ACMG/ AMP CC
Original ACMG/AMP Specification Stand Very rule summary alone strong
Strong
Moderate
Supporting
Comments
BP5
Variant found in a case with an na alternate molecular basis for
disease.
In rare circumstances, a patient can carry two variants in genes predisposing to hematologic malignancies.
BP6
Reputable source recently reported na variants as benign, but data are not available for laboratories to
perform independent evaluations.
According to SVI recommendations.
BP7
A synonymous variant for which General na na splicing prediction algorithms rec
predict no impact to the splice
consensus sequence nor the
na
na
Per original
Also applicable to intronic/non-coding variants at or beyond positions +7/-21 for which (1) SSF and MES predict either an increase in the canonical splice site score or a decrease of the canonical splice site score by no more than 10% and no putative cryptic splice sites are created AND (2) evolutionary conservation prediction algorithms predict the site as not conserved (e.g. PhyloP score <0.1 or the variant is the reference nucleotide in one primate and/or three mammal species.).
PVS1
Null variant in a gene for which LOF is a known mechanism
of disease.
Gene- na specific,
strength
Per modified RUNX1 PVS1 decision tree for SNV, indels and CNV and table of splicing effects.
na
RUNX1 LOF variants are a common mechanism of disease in FPD/AML. Three major isoforms (A, B, C) are expressed by use of two promotors and alternative splicing. C-terminal variants not predicted to undergo NMD are classified as PVS1_strong, deletions of exons 2 and 3, presumably only affecting RUNX1 isoform 1C,
PS1
Same AA change as a previously established pathogenic variant regardless of nucleotide change.
Strength
na na SameAA change as a previously established pathogenic
SameAA change as a previously established likely pathogenic variant
na
(1) RNA data or agreement in splicing predictors show no splicing effects (SSF and MES predict either increase in canonical splice site score or decrease of canonical splice score by no more than 10% and no putative splice site are created). (2) The previously established PATH/LPATH variant must be asserted pathogenic/likely pathogenic based on MM-VCEP rules for RUNX1 before this rule can be applied.
PS2
De novo (maternity and paternity confirmed) in a patient with the disease and no family history.
Disease- specific, strength
na na
na
nucleotide change. ≥ 2 proven
1 proven
(1) No family history is defined as: absence of the variant and any of the RUNX1-phenotypic criteria in first- and/or second-degree relatives. (2) The proband must exhibit at least one phenotypic FPD/AML criterion. (3) The
creation of a new splice site AND the nucleotide is not highly conserved.
ACMG/AMP guidelines. BP7 cannot be applied in combination with PP3.
variant regardless of nucleotide change.
regardless of
de novo
de novo
occurrences occurrence (maternity (maternity
site score or a decrease of the canonical splice site score by no more than 10%, and (3) no putative cryptic splice sites are created. BP4 should also be applied for synonymous, intronic and non-coding variants for which SSF and MES predict either an increase in the canonical splice site score or a decrease of the canonical splice site score by no more than 10% and no putative cryptic splice sites are created.
meet PVS1_moderate.
continued on the next page