Page 30 - Haematologica April 2020
P. 30
D. Wu et al.
872
haematologica | 2020; 105(4)
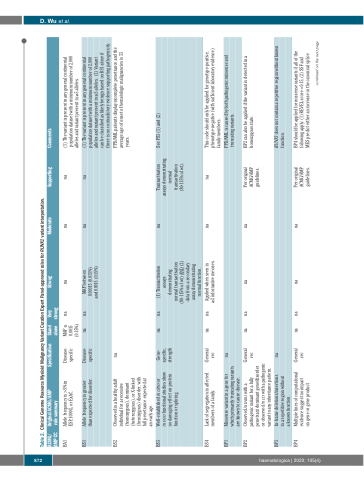
Table 2. Clinical Genome Resource Myeloid Malignancy Variant Curation Expert Panel-approved rules for RUNX1 variant interpretation.
ACMG/ AMP CC
Original ACMG/AMP rule summary
Specification
Stand alone
Very strong
Strong
Moderate
Supporting
Comments
BA1
Allele frequency is >5% in ESP, 1000G, or ExAC.
Disease- specific
MAF ≥ 0.0015 (0.15%)
na
na
na
na
(1) The variant is present in any general continental population dataset with a minimum number of 2,000
BS1
Allele frequency is greater than expected for disorder.
Disease- specific
na
na
MAF between 0.00015 (0.015%) and 0.0015 (0.15%)
na
na
alleles and variant present in ≥5 alleles.
(1) The variant is present in any general continental
BS2
Observed in a healthy adult individual for a recessive (homozygous), dominant (heterozygous), or X-linked (hemizygous) disorder, with full penetrance expected at an early age.
na
FPD/AML patients display incomplete penetrance and the average age of onset of hematologic malignancies is 33
BS3
Well-established in vitro or
in vivo functional studies show no damaging effect on protein function or splicing.
Gene- specific, strength
na
na
(1) Transactivation assays demonstrating normal transactivation (80-115% of wt) AND (2) data from a secondary assay demonstrating normal function.
na
Transactivation assays demonstrating normal transactivation (80-115% of wt).
See PS3 (1) and (2)
BS4
Lack of segregation in affected members of a family.
General rec
na
na
Applied when seen in ≥2 informative meioses.
na
na
This code should only be applied for genotype-positive, phenotype-negative (with sufficient laboratory evidence) family members.
BP1
Missense variant in a gene for which primarily truncating variants are known to cause disease.
na
FPD/AML is caused by both pathogenic missense and truncating variants.
BP2
Observed in trans with a pathogenic variant for a fully penetrant dominant gene/disorder or observed in cis with a pathogenic variant in any inheritance pattern.
General rec
na
na
na
na
Per original ACMG/AMP guidelines.
BP2 can also be applied if the variant is detected in a homozygous state.
BP3
In-frame deletions/insertions in a repetitive region without a known function.
na
RUNX1 does not contain a repetitive region without known function.
BP4
Multiple lines of computational evidence suggest no impact
on gene or gene product.
General rec
na
na
na
na
Per original ACMG/AMP guidelines.
BP4 should be applied for missense variants if all of the following apply: (1) REVEL score <0.15, (2) SSF and MES predict either an increase in the canonical splice
population dataset with a minimum number of 2,000
alleles and variant present in ≥5 alleles. (2) Variant
can be classified as likely benign based on BS1 alone if there is no contradictory evidence supporting pathogenicity.
years.
continued on the next page