Page 282 - Haematologica April 2020
P. 282
V. Daidone et al.
4A). Analyzing the intracellular content of the transfected cells revealed a similar picture, with rvWF totally absent in the homozygous condition, and 30% lower than normal vWF in the heterozygous condition (Figure 4B). The vWF:CB/vWF:Ag and vWF:FVIIIB/vWF:Ag ratios (respec- tively indicative of the multimer organization of vWF and its capacity for binding FVIII) did not change when RNAI was co-expressed with the WT, which means that no functional alterations are associated with P1 vWF (Figure 4A). These results demonstrate that the P1 mutation behaves like a gene null mutation, inducing a pure quanti- tative vWF defect.
RNAII and RNAIII - Transfection of HEK293T cells with pRc/CMV-vWF-RNAII caused a reduction in rvWF secre- tion of about 40% or 60%, respectively, if the vector was expressed with or without the WT counterpart (Figure 4A). Conversely, there was an increase in rvWF intracellu- lar content in the transfected cells that was more pro- nounced in the homozygous condition (Figure 4B), a find- ing suggestive of a partial retention of the P2 rvWF. A sim- ilar picture was seen for pRc/CMV-vWF-RNAIII: its expression was associated with a drop in rvWF secretion in the conditioned medium (of about 40% and 65% for the heterozygous and homozygous conditions, respec- tively), and a corresponding increase in the intracellular rvWF content (Figure 4B). The vWF:CB ratios were higher than normal, for both P2 and P3 rvWF (Figure 4A); in the homozygous condition, the ratios were 158.1% and
197.3%, respectively, of the WT (taken to be 100%), sug- gesting the presence of larger than normal vWF multimers. On the other hand, the vWF:FVIIIB ratios were lower when P2 and P3 were expressed at heterozygous level (i.e. 57.3% and 52.0% of the WT, respectively), and vWF:FVI- IIB was completely undetectable when each mutation was expressed at homozygous level. The P2 and P3 vWF muta- tions therefore both cause a partial quantitative defect associated with a defective FVIII binding capacity of the mutated vWF.
Multimer analysis on the rvWF in the conditioned media confirmed the above results: oligomers of unusually high molecular weight were apparent for both P2 and P3 rvWF, with an accumulation of vWF oligomers on the boundary between the stacking and running gel and a rel- ative representation of vWF multimers shifted towards the high-molecular-weight forms (Figure 5). All mutated rvWF oligomers also showed a delayed migration pattern, compared with WT (Figure 5), like the one seen in the patient’s platelet multimers. This picture points to the per- sistence of vWFpp, a condition associated with vWF hav- ing an impaired FVIII carrier capacity.28
Co-transfection with RNAI, RNAII and RNAIII - Co-trans- fecting HEK293T cells with equal amounts of all vectors resulted in a 60% reduction in secreted rvWF, with a cor- responding approximately 25% increase in intracellular rvWF content. In the conditioned medium, the rvWF:CB ratio was higher than normal, while vWF:FVIIIB levels
A
B
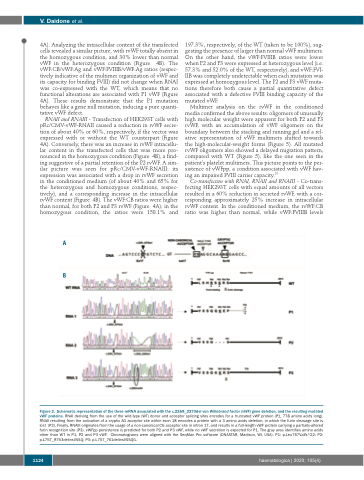
Figure 3. Schematic representation of the three mRNA associated with the c.2269_2270del von Willebrand factor (vWF) gene deletion, and the resulting mutated vWF proteins. RNAI deriving from the use of the wild-type (WT) donor and acceptor splicing sites encodes for a truncated vWF protein (P1, 778 amino acids long). RNAII resulting from the activation of a cryptic AG acceptor site within exon 18 encodes a protein with a 3 amino acids deletion, in which the furin cleavage site is lost (P2). Finally, RNAIII originates from the usage of a non-canonical CG acceptor site in intron 17, and results in a full-length vWF protein carrying a partially-altered furin recognition site (P3). vWFpp persistence is predicted for both P2 and P3 vWF, while no vWF secretion is expected for P1. The gray area identifies amino acids other than WT in P1, P2 and P3 vWF. Chromatograms were aligned with the SeqMan Pro software (DNASTAR, Madison, WI, USA). P1: p.Leu757Valfs*22; P2: p.L757_R763delinsVSSQ; P3: p.L757_761delinsVSSQG.
1124
haematologica | 2020; 105(4)