Page 274 - Haematologica April 2020
P. 274
L. Zheng et al.
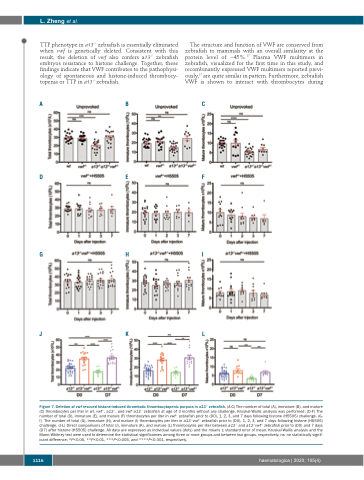
TTP phenotype in a13-/- zebrafish is essentially eliminated when vwf is genetically deleted. Consistent with this result, the deletion of vwf also confers a13-/- zebrafish embryos resistance to histone challenge. Together, these findings indicate that VWF contributes to the pathophysi- ology of spontaneous and histone-induced thrombocy- topenia or TTP in a13-/- zebrafish.
The structure and function of VWF are conserved from zebrafish to mammals with an overall similarity at the protein level of ~45%.17 Plasma VWF multimers in zebrafish, visualized for the first time in this study, and recombinantly expressed VWF multimers reported previ- ously,17 are quite similar in pattern. Furthermore, zebrafish VWF is shown to interact with thrombocytes during
ABC
DEF
GHI
JKL
Figure 7. Deletion of vwf rescued histone-induced thrombotic thrombocytopenic purpura in a13-/- zebrafish. (A-C) The number of total (A), immature (B), and mature (C) thrombocytes per liter in wt, vwf-/-, a13-/-, and vwf-/-a13-/- zebrafish at age of 3 months without any challenge. Kruskal-Wallis analysis was performed. (D-F) The number of total (D), immature (E), and mature (F) thrombocytes per liter in vwf-/- zebrafish prior to (D0), 1, 2, 3, and 7 days following histone (H5505) challenge. (G- I) The number of total (G), immature (H), and mature (I) thrombocytes per liter in a13-/-vwf-/- zebrafish prior to (D0), 1, 2, 3, and 7 days following histone (H5505) challenge. (J-L) Direct comparisons of total (J), immature (K), and mature (L) thrombocytes per liter between a13-/- and a13-/-vwf-/- zebrafish prior to (D0) and 7 days (D7) after histone (H5505) challenge. All data are expressed as individual values (dots) and the means ± standard error of mean. Kruskal-Wallis analysis and the Mann-Whitney test were used to determine the statistical significances among three or more groups and between two groups, respectively. ns: no statistically signif- icant difference; *P<0.05, **P<0.01, ***P<0.005, and ****P<0.001, respectively.
1116
haematologica | 2020; 105(4)