Page 116 - Haematologica April 2020
P. 116
C. Egan et al.
A
B
C
D
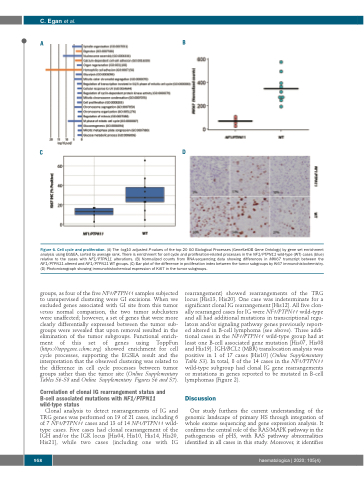
Figure 6. Cell cycle and proliferation. (A) The -log10 adjusted P-values of the top 20 GO Biological Processes (GeneSetDB Gene Ontology) by gene set enrichment analysis using EGSEA, sorted by average rank. There is enrichment for cell cycle and proliferation-related processes in the NF1/PTPN11 wild-type (WT) cases (blue) relative to the cases with NF1/PTPN11 alterations. (B) Normalized counts from RNA-sequencing data showing differences in MKI67 transcript between the NF1/PTPN11 altered and NF1/PTPN11 WT groups. (C) Bar plot of the difference in proliferation index between the tumor subgroups by Ki67 immunohistochemistry. (D) Photomicrograph showing immunohistochemical expression of Ki67 in the tumor subgroups.
groups, as four of the five NF1/PTPN11 samples subjected to unsupervised clustering were GI excisions. When we excluded genes associated with GI site from this tumor versus normal comparison, the two tumor subclusters were unaffected; however, a set of genes that were more clearly differentially expressed between the tumor sub- groups were revealed that upon removal resulted in the elimination of the tumor subgroups. Functional enrich- ment of this set of genes using ToppFun (https://toppgene.cchmc.org) showed enrichment for cell cycle processes, supporting the EGSEA result and the interpretation that the observed clustering was related to the difference in cell cycle processes between tumor groups rather than the tumor site (Online Supplementary Tables S6-S8 and Online Supplementary Figures S6 and S7).
Correlation of clonal IG rearrangement status and B-cell associated mutations with NF1/PTPN11 wild-type status
Clonal analysis to detect rearrangements of IG and TRG genes was performed on 19 of 21 cases, including 6 of 7 NF1/PTPN11 cases and 13 of 14 NF1/PTPN11 wild- type cases. Five cases had clonal rearrangement of the IGH and/or the IGK locus [His04, His10, His14, His20, His21], while two cases (including one with IG
rearrangement) showed rearrangements of the TRG locus [His13, His20]. One case was indeterminate for a significant clonal IG rearrangement [His12]. All five clon- ally rearranged cases for IG were NF1/PTPN11 wild-type and all had additional mutations in transcriptional regu- lators and/or signaling pathway genes previously report- ed altered in B-cell lymphoma (see above). Three addi- tional cases in the NF1/PTPN11 wild-type group had at least one B-cell associated gene mutation [His07, His08 and His19]. IGH/BCL2 (MBR) translocation analysis was positive in 1 of 17 cases [His10] (Online Supplementary Table S3). In total, 8 of the 14 cases in the NF1/PTPN11 wild-type subgroup had clonal IG gene rearrangements or mutations in genes reported to be mutated in B-cell lymphomas (Figure 2).
Discussion
Our study furthers the current understanding of the genomic landscape of primary HS through integration of whole exome sequencing and gene expression analysis. It confirms the central role of the RAS/MAPK pathway in the pathogenesis of pHS, with RAS pathway abnormalities identified in all cases in this study. Moreover, it identifies
958
haematologica | 2020; 105(4)