Page 114 - Haematologica April 2020
P. 114
C. Egan et al.
AB
CD
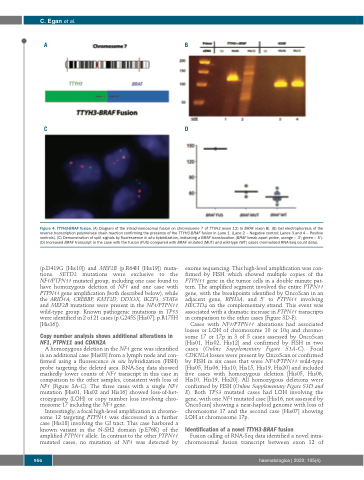
Figure 4. TTYH3-BRAF fusion. (A) Diagram of the intrachromosomal fusion on chromosome 7 of TTYH3 (exon 12) to BRAF (exon 8). (B) Gel electrophoresis of the reverse transcription polymerase chain reaction confirming the presence of the TTYH3-BRAF fusion in Lane 1. (Lane 2 – Negative control; Lanes 3 and 4 – Positive controls). (C) Demonstration of split signals by fluorescence in situ hybridization, indicating a BRAF translocation (BRAF break apart probe, orange – 3’; green – 5’). (D) Increased BRAF transcript in the case with the fusion (FUS) compared with BRAF mutated (MUT) and wild-type (WT) cases (normalized RNA-Seq count data).
(p.D419G [His10]) and MEF2B (p.R64H [His19]) muta- tions. SETD2 mutations were exclusive to the NF1/PTPN11 mutated group, including one case found to have homozygous deletion of NF1 and one case with PTPN11 gene amplification (both described below), while the ARID1A, CREBBP, KMT2D, DDX3X, IKZF3, STAT6 and MEF2B mutations were present in the NF1/PTPN11 wild-type group. Known pathogenic mutations in TP53 were identified in 2 of 21 cases (p.G245S [His07]; p.R175H [His16]).
Copy number analysis shows additional alterations in NF1, PTPN11 and CDKN2A
A homozygous deletion in the NF1 gene was identified in an additional case [His03] from a lymph node and con- firmed using a fluorescence in situ hybridization (FISH) probe targeting the deleted area. RNA-Seq data showed markedly lower counts of NF1 transcript in this case in comparison to the other samples, consistent with loss of NF1 (Figure 3A-C). The three cases with a single NF1 mutation [His01, His02 and His16] showed loss-of-het- erozygosity (LOH) or copy number loss involving chro- mosome 17 including the NF1 gene.
Interestingly, a focal high-level amplification in chromo- some 12 targeting PTPN11 was discovered in a further case [His18] involving the GI tract. This case harbored a known variant in the N-SH2 domain (p.E76K) of the amplified PTPN11 allele. In contrast to the other PTPN11 mutated cases, no mutation of NF1 was detected by
exome sequencing. This high-level amplification was con- firmed by FISH which showed multiple copies of the PTPN11 gene in the tumor cells in a double minute pat- tern. The amplified segment involved the entire PTPN11 gene, with the breakpoints identified by OncoScan in an adjacent gene, RPH3A, and 5’ to PTPN11 involving HECTD4 on the complementary strand. This event was associated with a dramatic increase in PTPN11 transcripts in comparison to the other cases (Figure 3D-F).
Cases with NF1/PTPN11 alterations had associated losses or LOH of chromosome 10 or 10q and chromo- some 17 or 17p in 3 of 5 cases assessed by OncoScan [His01, His02, His12] and confirmed by FISH in two cases (Online Supplementary Figure S3A-C). Focal CDKN2A losses were present by OncoScan or confirmed by FISH in six cases that were NF1/PTPN11 wild-type [His05, His06, His10, His15, His19, His20] and included five cases with homozygous deletion [His05, His06, His10, His19, His20]. All homozygous deletions were confirmed by FISH (Online Supplementary Figure S3D and E). Both TP53 mutated cases had LOH involving the gene, with one NF1 mutated case [His16, not assessed by OncoScan] showing a near-haploid genome with loss of chromosome 17 and the second case [His07] showing LOH at chromosome 17p.
Identification of a novel TTYH3-BRAF fusion
Fusion calling of RNA-Seq data identified a novel intra- chromosomal fusion transcript between exon 12 of
956
haematologica | 2020; 105(4)