Page 113 - Haematologica April 2020
P. 113
Molecular pathogenesis of histiocytic sarcoma
A
BC
DE
F
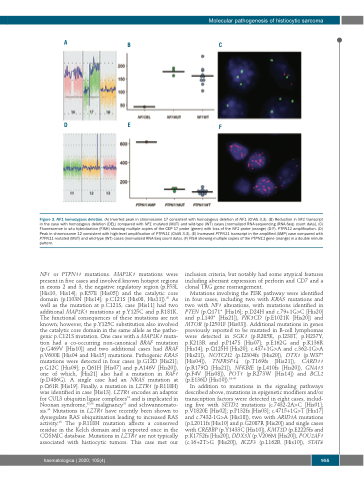
Figure 3. NF1 homozygous deletion. (A) Inverted peak in chromosome 17 consistent with homozygous deletion of NF1 (ChAS 3.3). (B) Reduction in NF1 transcript in the case with homozygous deletion (DEL) compared with NF1 mutated (MUT) and wild-type (WT) cases [normalized RNA-sequencing (RNA-Seq) count data]. (C) Fluorescence in situ hybridization (FISH) showing multiple copies of the CEP 17 probe (green) with loss of the NF1 probe (orange) (D-F). PTPN11 amplification. (D) Peak in chromosome 12 consistent with high-level amplification of PTPN11 (ChAS 3.3). (E) Increased PTPN11 transcript in the amplified (AMP) case compared with PTPN11 mutated (MUT) and wild-type (WT) cases (normalized RNA-Seq count data). (F) FISH showing multiple copies of the PTPN11 gene (orange) in a double minute pattern.
NF1 or PTPN11 mutations. MAP2K1 mutations were present in five cases and involved known hotspot regions in exons 2 and 3, the negative regulatory region (p.F53L [His10, His14]; p.K57E [His05]) and the catalytic core domain (p.I103N [His14]; p.C121S [His08, His11]).30 As well as the mutation at p.C121S, case [His11] had two additional MAP2K1 mutations at p.Y125C and p.R181K. The functional consequences of these mutations are not known; however, the p.Y125C substitution also involved the catalytic core domain in the same allele as the patho- genic p.C121S mutation. One case with a MAP2K1 muta- tion had a co-occurring non-canonical BRAF mutation (p.G469V [His10]) and two additional cases had BRAF p.V600E [His04 and His15] mutations. Pathogenic KRAS mutations were detected in four cases (p.G12D [His21]; p.G12C [His09]; p.Q61H [His07] and p.A146V [His20]), one of which, [His21] also had a mutation in RAF1 (p.D486G). A single case had an NRAS mutation at p.Q61R [His19]. Finally, a mutation in LZTR1 (p.R118H) was identified in case [His13]. LZTR1 encodes an adaptor for CUL3 ubiquitin ligase complexes31 and is implicated in Noonan syndrome,32,33 malignancy31 and schwannomato- sis.34 Mutations in LZTR1 have recently been shown to dysregulate RAS ubiquitination leading to increased RAS activity.35 The p.R118H mutation affects a conserved residue in the Kelch domain and is reported once in the COSMIC database. Mutations in LZTR1 are not typically associated with histiocytic tumors. This case met our
inclusion criteria, but notably had some atypical features including aberrant expression of perforin and CD7 and a clonal TRG gene rearrangement.
Mutations involving the PI3K pathway were identified in four cases, including two with KRAS mutations and two with NF1 alterations, with mutations identified in PTEN (p.Q171* [His16]; p.D24H and c.79+1G>C [His20] and p.L140* [His21]), PIK3CD (p.E1021K [His20]) and MTOR (p.I2501F [His03]). Additional mutations in genes previously reported to be mutated in B-cell lymphomas were detected in SGK1 (p.R285K, p.I238T, p.H237Y, p.K213R and p.P147S [His07]; p.E162G and p.K136R [His14]; p.Q125H [His20]; c.437+1G>A and c.362-1G>A [His21]), NOTCH2 (p.I2304fs [His20]), DTX1 (p.W37* [His04]), TNFRSF14 (p.T169fs [His21]), CARD11 (p.R179Q [His21]), NFKBIE (p.L410fs [His20]), GNA13 (p.F4V [His08]), POT1 (p.R273W [His14]) and BCL2 (p.E136D [His10]).36-40
In addition to mutations in the signaling pathways described above, mutations in epigenetic modifiers and/or transcription factors were detected in eight cases, includ- ing five with SETD2 mutations (c.7432-2A>C [His01]; p.V1820E [His02]; p.P132fs [His03]; c.4715+1G>T [His17] and c.7432-1G>A [His18]), two with ARID1A mutations (p.L2011fs [His10] and p.G2087R [His20]) and single cases with CREBBP (p.Y1433C [His10]), KMT2D (p.E2225fs and p.K1752fs [His20]), DDX3X (p.V206M [His20]), POU2AF1 (c.16+2T>G [His20]), IKZF3 (p.L162R [His10]), STAT6
haematologica | 2020; 105(4)
955