Page 111 - Haematologica April 2020
P. 111
Molecular pathogenesis of histiocytic sarcoma
and therefore predicted to be inactivating. The single mis- sense mutation (p.V1182D) was predicted to be deleterious or probably damaging by functional impact algorithms SIFT25 and PolyPhen-2.26 In addition to NF1 mutations, 3 of the 5 cases showed concurrent mutations in PTPN11 (p.F71V [His01]; p.E76G [His02]; p.A72V [His12]). PTPN11 mutations were present within the autoinhibitory N-SH2 domain at amino acid residues known to be associated with a gain-of-function consequence and described in Noonan syndrome and juvenile myelomonocytic leukemia
(JMML).27,28 One case without a PTPN11 mutation [His17] had an additional mutation in GNAI2 at p.R179H, a codon previously shown to be targeted by activating mutations29 and 1 of the 5 NF1 mutated cases also had a mutation in JAK2 at p.V617F [His12]. A fourth PTPN11 mutated case at p.E76K [His18] did not have another RAS pathway muta- tion; however, it had high level amplification of the mutat- ed PTPN11 allele (see below).
Additional mutations involving the RAS/MAPK path- way were detected in another 13 cases, none of which had
AB
CD
EF
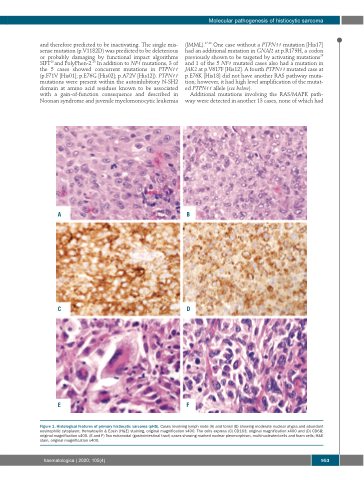
Figure 1. Histological features of primary histiocytic sarcoma (pHS). Cases involving lymph node (A) and tonsil (B) showing moderate nuclear atypia and abundant eosinophilic cytoplasm; Hematoxylin & Eosin (H&E) staining, original magnification x400. The cells express (C) CD163; original magnification x400 and (D) CD68; original magnification x400. (E and F) Two extranodal (gastrointestinal tract) cases showing marked nuclear pleomorphism, multinucleated cells and foam cells; H&E stain, original magnification x400.
haematologica | 2020; 105(4)
953