Page 62 - Haematologica March 2020
P. 62
C. Zhang et al.
AB
C
D
EF
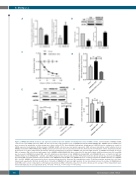
Figure 2. HMGB2 knockdown increases Lxn expression and decreases the number of hematopoietic stem cell (HSC) cell line. (A) Knockdown of HMGB2 in EML cells increases Lxn mRNA expression. EML cells were infected by control lentivirus (Con) or HMGB2 knockdown shRNA (HMGB2 KD). HMGB2 and Lxn mRNA levels were measured by quantitative real-time polymerase chain reaction (PCR). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was the endogenous control for mRNA expression normalization. (B) Knockdown of HMGB2 in EML cells increases Lxn protein expression. EML cells were infected by control lentivirus (Con) or HMGB2 knockdown shRNA (HMGB2 KD). HMGB2 and Lxn protein levels were measured by western blot. Actin was the normalization control. (Top) Representative western blot out of three independent experiments. (Bottom) Quantification of intensity of HMGB2 (left) and Lxn (right) proteins. (C) HMGB2 knockdown decreases EML cell number. EML cells infected with empty (Con) or HMGB2 shRNA (HMGB2 KD) were cultured for 12 days and counted at different time points. (D) Lxn mRNA was decreased in HMGB2-knockdown EML cells with simultaneous knockdown of Lxn. HMGB2-knockdown EML cells (HMGB2 KD) were co-transfected with Lxn shRNA lentiviral vector (HMGB2 KD + Lxn KD). Lxn mRNA and protein was measured by real-time PCR and western blot. (E) Lxn protein was decreased in HMGB2- knockdown EML cells with simultaneous knockdown of Lxn. HMGB2-knockdown EML cells (HMGB2 KD) were co-transfected with Lxn shRNA lentiviral vector (HMGB2 KD + Lxn KD). HMGB2 and Lxn protein levels were measured by western blot. Actin was the normalization control. (Top) Representative western blot out of three independent experiments. (Bottom) Quantification of intensity of HMGB2 (left) and Lxn (right) proteins. (F) Lxn knockdown restores the number of HMGB2-knock- down EML cells to a level comparable to control group. Data shown are EML cell number at day 3 of cell culture. All data are the average of three independent exper- iments with triplicates in each experiment (n=9). *P<0.05; **P<0.01; ***P<0.001; ****P<0.0001.
576
haematologica | 2020; 105(3)