Page 60 - Haematologica March 2020
P. 60
C. Zhang et al.
to stress or injury. The flexibility of stem cells to adapt to physiological needs is achieved by precise regulation of self-renewal and differentiation. Many molecules and sig- naling pathways have been found to be involved in this process.3 However, identification of the collection of genes contributing to critical stem cell functions is far from com- plete.
Hematopoietic stem cell number and function exhibit natural variation among humans as well as among differ- ent mouse strains.5-8 The natural variation is largely attrib- uted to DNA variants in the genome that function as reg- ulatory elements to control gene expression.9 The genetic
A
BC
diversity is a powerful but underused tool for unraveling the critical gene networks in stem cell regulation. Using genome-wide association studies, increasing numbers of gene regulatory variants have been strongly implicated in hematologic phenotypes and diseases in humans, such as fetal hemoglobin-associated genetic variants in patients with sickle cell disease (SCD) and β-thalassemia.10,11 Recently, several reports have revealed an important yet previously unrecognized role of genetic variants in regu- lating epigenetics.9,12-15 For example, DNA variations may affect the recruitment and binding affinity of transcription factors, which in turn lead to histone tail modifications.
DE
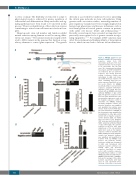
Figure 1. HMGB2 suppresses Lxn promoter activity. (A) Lxn promoter
sequence spans from nucleotides upstream of the tran- scription start site (+1) of Lxn gene to 27 nucleotides into the first exon. The chromosomal positions for Lxn gene and SNP rs31528793 are indicated. (B) Lxn promoter sequence has strong promoter activity. Luciferase activity was determined in HEK cells trans- duced with luciferase reporter con- struct containing either Lxn pro- moter sequence (Lxn-PGL3) or con- trol vector (PGL3). (C) HMGB2 specifically binds to Lxn promoter sequence. Cromatin immunopre- cipitation (ChIP) assay was per- formed with an HMGB2 polyclonal antibody (HMGB2) or IgG control (IgG). The genomic sequence in the 500 base pairs downstream of the Lxn promoter region were used as the negative sequence control to determine the HMGB2 binding specificity (Negative control). Lxn promoter sequence was amplified and quantified by real-time poly- merase chain reaction (PCR) (top). The fold enrichment of HMGB2 in the Lxn promoter was quantified by normalization to either IgG control (bottom left) or negative sequence control (bottom right). (D) H2A.X does not bind to Lxn promoter. ChIP assay was performed with an H2A.X polyclonal antibody (H2A.X) or IgG control (IgG). The genomic sequence in the 500 base pairs downstream of the Lxn promoter region were used as the negative sequence control to determine the H2A.X binding specificity (Negative control). Lxn promoter sequence was amplified and quantified by real-time PCR. (E) HMGB2 sup- presses Lxn promoter activity. Luciferase activity was determined in HEK cells transduced with luciferase reporter construct con-
taining either Lxn
sequence (Lxn-PGL3) or control vector with scramble sequence (Scramble-PGL3) without or with HMGB2 plasmid (Scramble-PGL3 + HMGB2, and Lxn-PGL3 + HMGB2). Data are the average of three inde- pendent experiments with tripli- cates in each experiment (n=9). *P<0.05; **P<0.01; ***P<0.001.
promoter
333
574
haematologica | 2020; 105(3)