Page 99 - 2020_02-Haematologica-web
P. 99
Irf2bp2b regulates zebrafish NMP cell fate choice
strated that zebrafish Irf2bp2b inhibits pu.1 expression. Thus, we investigated the reason underlying the repres- sive property of IRF2BP2.
Post-translational modification of proteins plays a piv- otal role in regulating their function. SUMOylation is an important type of post-translational modification which involves a cascade of dedicated enzymes that facilitate the covalent modification of specific lysine residues on target proteins with monomers or polymers of SUMO (small ubiquitin-like modifier).38 The SUMOylation of substrate proteins is frequently linked with transcriptional repres- sion.39 In fact, multiple adducts (the smallest one was about 10 kD larger than the unmodified protein, which was nearly the size of one SUMO molecule) of Irf2bp2b were detected by western blot (Figure 7A). The SUMO- targeted lysine usually lies in the canonical motif Ψkxe.40 A SUMO consensus motif VKKE (lysine 496) located at the C-terminus of Irf2bp2b was predicted by bioinformat-
ics (Online Supplementary Figure S1). The putative lysine was mutated to arginine (Irf2bp2bK496R) to abolish covalent binding with the SUMO molecule. The modified bands of the Irf2bp2bK496R mutant protein disappeared as expected (Figure 7A, B). In addition, an Irf2bp2bE498A mutant was constructed to destroy the conservation of the SUMO consensus motif which still allowed the accessibility of lysine 496 to other modifiers. The modified bands disap- peared as Irf2bp2bK496R mutant did (Figure 7C), indicating that Irf2bp2b is a SUMOylated substrate.
In HEK293T cells, GFP-SUMO was co-transfected with HA-tagged wildtype Irf2bp2b or Irf2bp2bK496R mutant. Immunoprecipitation assays showed that GFP-SUMO co- precipitated with HA-tagged wildtype Irf2bp2b, but not with the Irf2bp2bK496R mutant (Figure 7D). This further indicated that Irf2bp2b is indeed SUMOylated in cells.
Luciferase reporter assays with zebrafish pu.1 promoter were then conducted to assess the repressive capacity of
ABC
D
K
EF
G
I
H
J
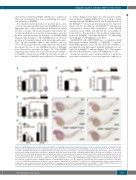
Figure 6. DNA-binding property is indispensable for Irf2bp2b in regulating neutrophil-macrophage progenitor cell fate (continued). (A) Ability of Irf2bp2b mutants to repress the zebrafish pu.1 promoter (-1.7kb). Error bars represent the mean ± standard error of mean (SEM) of at least three replicates. ***P<0.001 [analysis of variance (ANOVA) followed by the least significant difference (LSD) post-hoc test for multiple comparisons]. (B) Irf2bp2b represses luciferase expression from the pu.1 promoter 132bp A region (from -1308 bp to -1439 bp). Error bars represent the mean ± standard deviation (SD) of at least three replicates. ***P<0.001 (Student t test). (C) Chromatin immunoprecipitation polymerase chain reaction analysis of pu.1 promoter A region in zebrafish embryos expressing green fluorescent protein (GFP), Irf2bp2b-GFP or Irf2bp2bR55/59L-GFP using an anti-GFP antibody. The position of the primers used to amplify the pu.1 promoter A region are indicated with red arrows. Error bars represent the mean ± SEM of at least three replicates. ****P<0.0001 (ANOVA followed by the LSD post-hoc test for multiple comparisons). (D) Luciferase repression assays of Irf2bp2b mutants on zebrafish pu.1 promoter (-1.7 kb). Error bars represent the mean ± SEM of at least three replicates. ***P<0.001 (ANOVA followed by the LSD post-hoc test for multiple comparisons). (E-J) Irf2bp2bR55/59L mRNA rescue assays in irf2bp2b-/- mutant embryos. Mpx and mfap4 probes were used in whole-mount in situ hybridization to examine rescue effects associated with injection of Irf2bp2bR55/59L mutant mRNA. (K) Error bars rep- resent the mean ± SEM of at least three replicates. ***P<0.001 (ANOVA followed by the LSD post-hoc test for multiple comparisons).
haematologica | 2020; 105(2)
333