Page 137 - 2019_12-Haematologica-web
P. 137
Single-agent CHK1 inhibition in CLL
A
B
CD
E
F
G
H
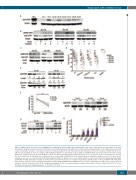
Figure 5. CHK1 protein level and effects of MU380 in non-stimulated chronic lymphocytic leukemia (CLL) cells. (A) The CHK1 protein was detectable in all tested CLL samples using the sensitive detection kit. (B) The treatment with fludarabine [10 μM; 24 hours (h)] resulted in phosphorylation of the CHK1 protein on Ser345 residue indicating its activation. (C) Reduction of pS296 autophosphorylation after MU380 treatment (400 nM, 24 h). (D) The 72 h treatment with MU380 (100-400 nM) decreased viability of most CLL samples, with insignificant differences among the studied samples; wt-ATM/wt-TP53 (wt) versus TP53-mut P=0.199; versus ATM-mut P=0.964; versus 11q- (the other ATM allele intact) P=0.849. The healthy peripheral blood mononuclear cell samples (n=3) were substantially less affected (P<0.001). (E) MU380 elicited apoptosis as evidenced by the cleaved PARP (C-PARP) protein. The values indicate densitometric analysis set to 1.0 in control. (F, left) Viability decrease in CLL cells transfected with siRNA targeting CHEK1. (F, right) Decrease in the CHK1 protein level after transfection with siRNA targeting CHEK1. (G) MU380 (400 nM; 24 h) did not change the p53 protein level in TP53-wt samples, in contrast to fludarabine (10 μM; positive control). (H) MU380 (400 nM; 24 h) did not induce expression of p53-downstream target genes BAX, PUMA, GADD45A, and CDKN1A (p21), in contrast to fludarabine (10 μM; positive control). The fold change is related to untreated control (CTR). The graph summarizes results of real-time polymerase chain reaction analyses in three samples (CLL-58, CLL-77, CLL- 83). Error bars represent standard deviation. ***P<0.001; **P<0.01.
haematologica | 2019; 104(12)
2451