Page 114 - 2019_11 Resto del Mondo-web
P. 114
C. Di Genua et al.
expression of Gja1 in AKM HSC may lead to loss of p53. Analysis of biological GO terms showed a number of metabolic processes up-regulated in AKM versus AM HSC (Online Supplementary Table S4) indicating metabolic dys- regulation could also be involved in loss of quiescence in AKM HSC.38
Finally, in order to determine relevance of the genes/pathways described for human AML1-ETO, we studied genes that are up-regulated in human HSC trans- duced with AML1-ETO.39 There was an enrichment of these genes in AM HSC compared to AKM, suggest a cor- relation between human and mouse AML1-ETO target genes (Figure 5K).
Collectively, these results demonstrate that K-RasG12D is detrimental to HSC harboring Aml1-ETO, causing a loss
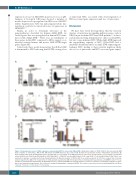
AB
of functional HSC, associated with down-regulation of HSC-associated gene expression and loss of quiescence.
Discussion
We have here tested the hypothesis that the observed absence of mutations in signaling pathway genes, such as KRAS, in pre-leukemic HSC from AML patients11,12 is due to such mutations being detrimental not only to normal HSC, but also to pre-leukemic HSC. While Aml1-ETO improved the repopulating capacity of HSC, K-RasG12D had a markedly detrimental effect on Aml1-ETO-expressing pre- leukemic HSC, leading to their eventual depletion, likely due to loss of quiescence and HSC-associated gene expres-
C
DE
Figure 3. Hematopoietic stem cell (HSC) expansion caused by Aml1-ETO is reversed by K-RasG12D. (A) Absolute number of CD45.2 HSC in the bone marrow (BM) from recipients of CON (n=11), AM (n=14), KM (n=13) and AKM fetal liver (FL) cells (n=14). Results were generated in three independent experiments; (B) Representative FACS plots showing gating used to quantify HSC as a percentage of the BM mononuclear cells across all experiments. (C) Percentage reconstitution of total CD45.2 cells, CD45.2 myeloid (LiveCD19–CD4–CD8a–NK1.1–), CD45.2 B cells (LiveNK1.1-Mac1-CD19+) and CD45.2 T cell (LiveNK1.1–Mac1–CD4+CD8a+) com- partments in primary, secondary and tertiary transplantations. (D) Absolute number of CD45.2 HSC in secondary recipients of CON (n=9 recipient mice in 2 inde- pendent experiments), AM (n=10 recipient mice in 3 independent experiments), KM (n=4 recipient mice in 2 independent experiments), and AKM FL cells (n=7 recip- ient mice in 2 independent experiments). (E) Replating efficiency of CD45.2 LSK BM cells. Average number of colonies is shown for 5-6 biological replicates per geno- type in two independent experiments. The results were analyzed using multiple comparison ANOVA. The results are presented as the mean±Standard Error of Mean. *P<0.05; **P<0.01; ***P<0.001; ****P<0.0001.
2220
haematologica | 2019; 104(11)