Page 235 - Haematologica Vol. 107 - September 2022
P. 235
LETTER TO THE EDITOR
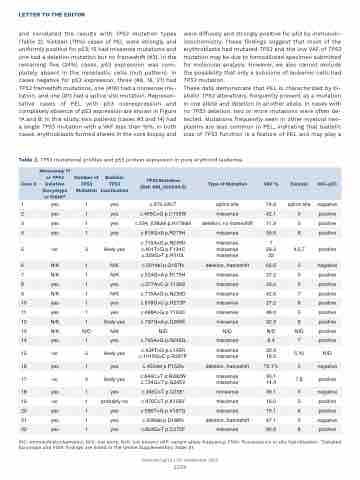
and correlated the results with TP53 mutation types (Table 2). Sixteen (76%) cases of PEL were strongly and uniformly positive for p53; 15 had missense mutations and one had a deletion mutation but no frameshift (#3). In the remaining five (24%) cases, p53 expression was com- pletely absent in the neoplastic cells (null pattern). In cases negative for p53 expression, three (#6, 16, 21) had TP53 frameshift mutations, one (#18) had a nonsense mu- tation, and one (#1) had a splice site mutation. Represen- tative cases of PEL with p53 overexpression and completely absence of p53 expression are shown in Figure 1A and B. In this study, two patients (cases #3 and 14) had a single TP53 mutation with a VAF less than 15%. In both cases, erythroblasts formed sheets in the core biopsy and
were diffusely and strongly positive for p53 by immunoh- istochemistry. These findings suggest that most of the erythroblasts had mutated TP53 and the low VAF of TP53 mutation may be due to hemodiluted specimen submitted for molecular analysis. However, we also cannot exclude the possibility that only a subclone of leukemic cells had TP53 mutation.
These data demonstrate that PEL is characterized by bi- allelic TP53 alterations, frequently present as a mutation in one allele and deletion in another allele. In cases with no TP53 deletion, two or more mutations were often de- tected. Mutations frequently seen in other myeloid neo- plasms are less common in PEL, indicating that biallelic loss of TP53 function is a feature of PEL and may play a
Table 2. TP53 mutational profiles and p53 protein expression in pure erythroid leukemia.
Case #
Monosomy 17 or TP53 Deletion (karyotype or FISH)*
Number of
TP53
Mutation
Biallelic
TP53
Inactivation
TP53 Mutation (Ref: NM_000546.5)
Type of Mutation
VAF %
Exon(s)
IHC-p53
1
yes
1
yes
c.673-2A>T
splice site
74.9
splice site
negative
2
yes
1
yes
c.405C>G p.C135W
missense
42.1
5
positive
3
yes
1
yes
c.534_536del p.H179del
deletion, no frameshift
11.9
5
positive
4
yes
1
yes
c.818G>A p.R273H
missense
59.6
8
positive
5
no
3
likely yes
c.715A>G p.N239D c.401T>G p.F134C c.329G>T p.R110L
missense missense missense
1 29.3 32
4,5,7
positive
6
N/K
1
N/K
c.501del p.Q167fs
deletion, frameshift
62.6
5
negative
7
N/K
1
N/K
c.524G>A p.R175H
missense
37.2
5
positive
8
yes
1
yes
c.377A>C p.Y126S
missense
23.0
5
positive
9
N/K
1
N/K
c.715A>G p.N239D
missense
42.6
7
positive
10
yes
1
yes
c.818G>C p.R273P
missense
27.2
8
positive
11
yes
1
yes
c.488A>G p.Y163C
missense
48.0
5
positive
12
N/K
1
likely yes
c.797G>A p.G266E
missense
92.3
8
positive
13
N/K
N/D
N/K
N/D
N/D
N/D
N/D
positive
14
yes
1
yes
c.745A>G p.R249G
missense
8.4
7
positive
15
no
2
likely yes
c.434T>G p.L145R c.1010G>C p.R337P
missense missense
20.4 18.5
5,10
N/D
16
yes
1
yes
c.455del p.P152fs
deletion, frameshift
70.1%
5
negative
17
no
2
likely yes
c.844C>T p.R282W c.734G>T p.G245V
missense missense
35.1 14.4
7,8
positive
18
yes
1
yes
c.493C>T p.Q165*
nonsense
39.1
5
negative
19
no
1
probably no
c.476C>T p.A159V
missense
16.0
5
positive
20
yes
1
yes
c.590T>G p.V197G
missense
15.1
6
positive
21
yes
1
yes
c.558del p.D186fs
deletion, frameshift
47.1
5
negative
22
yes
1
yes
c.824G>T p.C275F
missense
80.6
8
positive
IHC: immunohistochemistry; N/D: not done; N/K: not known; VAF: variant allele frequency; FISH: fluorescence in situ hybridization. *Detailed karyotype and FISH findings are listed in the Online Supplementary Table S1.
Haematologica | 107 September 2022
2234