Page 61 - Haematologica May 2022
P. 61
FGG mutation in congenital afibrinogenemia
A
BC
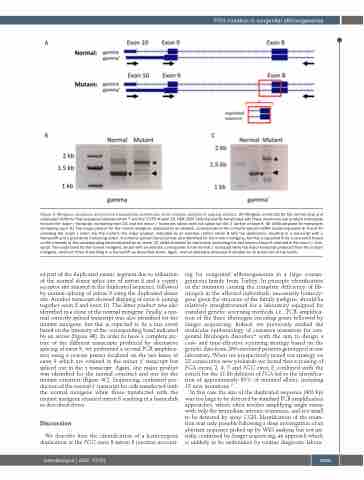
Figure 4. Minigene constructs and reverse transcription polymerase chain reaction analysis of splicing variants. (A) Minigene constructs for the normal (top) and duplicated (bottom) FGG sequence between intron 7 and the 3’UTR of exon 10. HEK-293T cells transiently transfected with these constructs can produce transcripts for both the major g transcript, containing exon 10, and the minor g’ transcript, which does not splice out the 3’ portion of exon 9. (B) cDNA obtained for transcripts containing exon 10. The major product for the normal minigene, indicated by an asterisk, corresponds to the correctly spliced mRNA containing exons 8, 9 and 10 encoding the major g chain. For the mutant, the major product, indicated by an asterisk, retains intron 8 with the duplication, resulting in a transcript with a frameshift and a premature truncating codon. A correctly spliced transcript was also identified for the mutant minigene, but this is expected to be a rare event based on the intensity of the corresponding band indicated by an arrow. (C) cDNA obtained for transcripts containing the last bases of exon 9 retained in the minor g’ tran- script. The major band for the normal minigene, shown with an asterisk, corresponds to the normal g’ transcript while the major transcript produced from the mutant minigene, retained intron 8 resulting in a frameshift as described above. Again, normal splicing is observed (indicated by an arrow) but at low levels.
ed part of the duplicated exonic segment due to utilisation of the normal donor splice site of intron 8 and a cryptic acceptor site situated in the duplicated sequence, followed by normal splicing of intron 9 using the duplicated donor site. Another transcript showed skipping of exon 9, joining together exon 8 and exon 10. The latter product was also identified in a clone of the normal minigene. Finally, a nor- mal correctly spliced transcript was also identified for the mutant minigene, but this is expected to be a rare event based on the intensity of the corresponding band indicated by an arrow (Figure 4B). In order to have a complete pic- ture of the different transcripts produced by alternative splicing of exon 9, we performed a second PCR amplifica- tion using a reverse primer localized on the last bases of exon 9 which are retained in the minor g’ transcript but spliced out in the g transcript. Again, one major product was identified for the normal construct and one for the mutant construct (Figure 4C). Sequencing confirmed pro- duction of the normal g’ transcript for cells transfected with the normal minigene while those transfected with the mutant minigene retained intron 8 resulting in a frameshift as described above.
Discussion
We describe here the identification of a homozygous duplication at the FGG exon 8-intron 8 junction account-
ing for congenital afibrinogenemia in a large consan- guineous family from Turkey. In principle identification of the mutation causing the complete deficiency of fib- rinogen in the affected individuals, necessarily homozy- gous given the structure of the family pedigree, should be relatively straightforward for a laboratory equipped for standard genetic screening methods i.e., PCR amplifica- tion of the three fibrinogen encoding genes followed by Sanger sequencing. Indeed, we previously studied the molecular epidemiology of causative mutations for con- genital fibrinogen disorders22 with the aim to design a cost- and time-effective screening strategy based on the genetic data from 266 unrelated patients genotyped in our laboratory. When we prospectively tested our strategy on 32 consecutive new probands we found that screening of FGA exons 2, 4, 5 and FGG exon 8 combined with the search for the 11 kb deletion of FGA led to the identifica- tion of approximately 80% of mutated alleles, including 15 new mutations.22
In this case the size of the duplicated sequence (403 bp) was too large to be detected by standard PCR amplification approaches, which often involve amplifying single exons with only the immediate intronic sequences, and too small to be detected by array CGH. Identification of the muta- tion was only possible following a deep investigation of an aberrant sequence picked up by WES analysis but not ini- tially confirmed by Sanger sequencing, an approach which is unlikely to be undertaken by routine diagnostic labora-
haematologica | 2022; 107(5)
1069