Page 116 - Haematologica May 2022
P. 116
J. Gao et al.
A
B
C
D
E
F
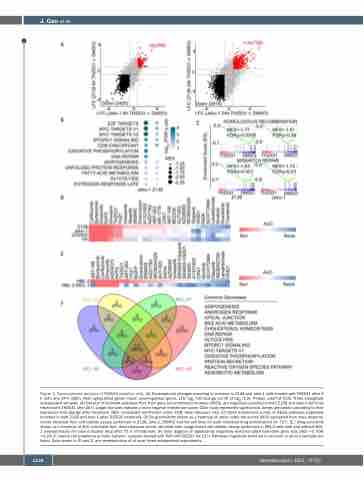
Figure 3. Transcriptomic analysis of THZ531-sensitive cells. (A) Transcriptome changes occurring in common in Z138 and Jeko-1 cells treated with THZ531 after 6 h (left) and 24 h (right). Red: upregulated genes, black: downregulated genes. LFC: log2 fold change cut-off of log2 (1.5), P-value cutoff of 0.05. Three biologically independent samples. (B) Dot plot of hallmark pathways that, from gene set enrichment analysis (GSEA), are negatively enriched in both Z138 and Jeko-1 cell lines treated with THZ531 after 24 h. Larger dot sizes indicate a more negative enrichment score. Color scale represents significance. Genes are ranked according to their expression fold change after treatment. NES: normalized enrichment score; FDR: false discovery rate. (C) GSEA enrichment curves of KEGG pathways negatively enriched in both Z138 and Jeko-1 after THZ531 treatment. (D) Drug sensitivity shown as a heatmap of areas under the curves (AUC) calculated from dose-response curves obtained from cell viability assays performed in Z138, Jeko-1, DOHH2 and Val cell lines for each indicated drug administered for 72 h. (E ) Drug sensitivity shown as a heatmap of AUC calculated from dose-response curves obtained from image-based cell viability assays performed in HBL-2 cells with and without MCL- 1 overexpression for each indicated drug after 72 h of treatment. (F) Venn diagram of significantly negatively enriched GSEA hallmarks gene sets (NES <0, FDR <0.25) in mantle cell lymphoma primary patients’ samples treated with 500 nM THZ531 for 12 h. Pathways negatively enriched in common in all four samples are listed. Data shown in (D and E) are representative of at least three independent experiments.
1124
haematologica | 2022; 107(5)