Page 138 - 2022_03-Haematologica-web
P. 138
M.C.J. Ma et al.
ABC
D
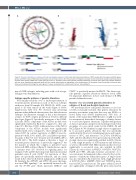
Figure 2. Structural alterations in subtypes of B-cell non-Hodgkin lymphoma. (A) A circos plot shows translocations of MYC (purple), BCL2 (orange) and BCL6 (green) genes, and GISTIC tracks of DNA copy number gains (red) and losses (blue). (B, C) Volcano plots of integrative analysis results showing the changes in gene expression of genes within peaks of DNA copy number gain (B) or loss (C). A positive T-test score indicates increased expression in tumors with a given copy number alteration, and vice versa. Significantly expressed genes with the correct directionality are highlighted in the shaded areas. (D) Oncoplots show the overlap of structural alter- ations and mutations that target the same genes. P-values are derived from a Fisher exact test (ns: not significant).
major B-NHL subtypes, including genes with a role in regu- lating protein ubiquitination.
Subtype-specific patterns of genetic alterations
We formally tested the over- or under-representation of recurrent genetic alterations in each of the four subtypes with more than 100 samples (BL, DLBCL, FL, MCL), com- pared to all other tumors in the study (Figure 4, Online Supplementary Table S13). We observed some interesting patterns within hallmark characteristics that differ between subtypes. An illustrative example of this is the alternative avenues for BAF complex perturbation between different histologies (Figure 5). Specifically, mutations of the SMAR- CA4 (aka. BRG1) component of the ATPase module were significantly enriched in BL (24%) compared to other sub- types (4%, Q-value <0.001), while mutations of the BCL7A component of the ATPase module were significantly enriched in FL (11%) compared to other subtypes (4%, Q- value=0.007). In contrast, mutations of ARID1A were fre- quent in both BL (19%) and FL (15%), and DNA copy num- ber gains of BCL11A were frequent in both DLBCL (28%) and FL (22%). The BAF complex is therefore a target of recurrent genetic alterations, as previously suggested,43 but the manner in which this complex is perturbed varies between B-NHL subtypes (Figure 5). Similar disease-specif- ic patterns were also observed for signaling genes; for example, TCF3 and ID3 have important functions in normal germinal center B cells (GCB),44 but mutations of these genes are specifically enriched within BL and are rarely found in the other GCB-derived malignancies, DLBCL and FL. Similarly, the ATP6AP1 and ATP6V1B2 genes that func- tion in mTOR signaling45,46 are specifically mutated in FL, and the DUSP2 gene which inactivates ERK1/247 and
STAT348 is specifically mutated in DLBCL. The disease-spe- cific patterns of genetic alterations therefore reveal subtle but important differences in how each subtype of B-NHL perturbs hallmark features.
Clusters of co-associated genomic alterations in subtypes of B-cell non-Hodgkin lymphoma
We next defined how each genetic alteration co-associat- ed with or mutually excluded other genetic alterations by pairwise assessments using false-discovery rate (FDR)-cor- rected Fisher tests (Online Supplementary Table S14). A matrix of the transformed FDR Q-values (-logQ) was used for unsupervised hierarchical clustering to identify clusters of co-associated genetic alterations. Together with patterns of disease-specificity, unsupervised clustering revealed clear groupings of co-associated events for BL, DLBCL, FL and MCL (Figure 4). We identified a single cluster of significant- ly co-associated genetic alterations that was specifically enriched in BL (Cluster 1), including mutations and translo- cations of MYC, and mutations of CCND3, SMARCA4, TCF3 and ID3 which have been previously reported in BL.4 A single cluster was significantly enriched in MCL (Cluster 7), with a high frequency of ATM mutations and deletions, as well as other DNA CNA. Other mutations that were not significantly co-associated were also enriched in MCL (Cluster 6), such as those in WHSC1, NOTCH1, NOTCH2, BCOR and UBR5, although statistical assessment of co- association may be hampered in this context by the low fre- quencies of mutations within these genes. A single cluster was also enriched in FL (Cluster 4), with a high prevalence of KMT2D, BCL2, CREBBP, EZH2 and TNFRSF14 muta- tions and BCL2 translocations. The genes within Cluster 4 also significantly overlapped with the previously reported
694
haematologica | 2022; 107(3)