Page 77 - 2022_02-Haematologica-web
P. 77
RXRA active mutation
AB
CD
EFG
H
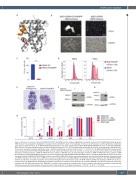
Figure 1. Monocytic maturation induced by RXRA DT448/9PP. (A) DT448/9 amino acids are highlighted in red within the structure of the RXRA ligand binding domain (PDB 4K4J). An Ncoa2 peptide is highlighted in orange. Note that DT448/9 occurs at the beginning of helix 12, the AF2 domain, and the rigid proline substitution could enhance helix formation. (B, C) RXR-KO KMT2A-MLLT3 leukemia cells were transduced with MSCV-RXRA DT448/9PP-IRES-mCherry or with MSCV-RXRA WT- IRES-mCherry. Cells were evaluated under fluorescent and light microscopy at 72 h. (B) Transduced cells were sorted for mCherry+ cells and plated in methylcellulose, and colonies assessed in technical triplicates on day 7. Statistical significance evaluated using the t-test, ***P<0.001 (C). (D) RXR-KO KMT2A-MLLT3 leukemia cells were transduced with MSCV-RXRA DT448/9PP-IRES-mCherry, stained with FxCycle violet, and retention of the dye was assessed by flow cytometry at the indicated time points comparing mCherry+ (RXRA DT 448/9 PP) versus mCherry– (RXR-KO) cells. (E) Cytomorphology of mCherry+ (RXRA DT 448/9 PP) versus mCherry– (RXRA WT endogenous) sorted KMT2A-MLLT3 leukemia cells after transduction with MSCV-RXRA DT448/9PP-IRES-mCherry. (F) RXR-KO KMT2A-MLLT3 leukemia cells were transduced with MSCV-RXRA DT448/9PP-IRES-mCherry or with MSCV-RXRA-IRES-mCherry and protein expression was evaluated through western blot analysis using anti-RXRA antibody (H-10, Santa Cruz). GAPDH was used as a loading control. (G) KMT2A-MLLT3 WT leukemia cells were transduced with MSCV-RXRA WT-IRES- mCherry as indicated, and protein expression was evaluated through western blot analysis using anti-RXRA antibody (5388, Cell Signaling). GAPDH was used as a loading control. (H) KMT2A-MLLT3 WT leukemia cells were transduced as indicated and analyzed by flow cytometry. RXRA WT cells were analyzed with and without treatment with 250 nM bexarotene for 24 h. RAW 264.7 cells were used as a positive staining control, but not included in the statistical comparisons. Statistical sig- nificance was evaluated using analysis of variance with the Tukey correction for multiple comparisons, *P<0.05. **P<0.01. ***P<0.001.
haematologica | 2022; 107(2)
419