Page 212 - 2022_01-Haematologica-web
P. 212
R.J. Leeman-Neill et al.
ABC
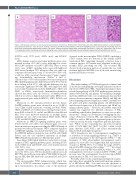
Figure 1. Morphologic spectrum of post-transplant plasmablastic lymphomas. Representative hematoxylin and eosin (H&E)-stained sections of post-transplant plasmablastic lymphomas - from (A) case 4, showing a monotonous infiltrate of plasmablasts, with insets highlighting areas of plasmablastic morphology (top) and focal areas of plasmacytic differentiation (bottom), (B) case 5, showing numerous tingible body macrophages that impart a “starry sky” appearance, and (C) case 7, showing pleomorphic morphology, with insets displaying areas of plasmablastic morphology (top) and anaplastic-appearing multinucleated cells (bottom).
KMT2A, n=2), TET2 (n=3), ASXL1 (n=2), and KDM5C (n=2).
DNA damage response and repair pathway genes were mutated in seven of 11 (64%) cases, including four of five (80%) EBV– and three of six (50%) EBV+ PBL. Three of seven (43%) cases, all EBV–, including both casese with high-level MSI, showed multiple TP53 mutations. One case with con- comitant subclonal monosomy 17 showed 30% P53+ cells, as did two with concurrent chromosome 17 gains, suggest- ing bi/multiallelic inactivation (Tables 2 and 3). Chromosome 17/TP53 abnormalities were more frequent in PBL exhibiting plasmacytic differentiation (4/5, 80%) than in those lacking it (2/6, 33%), although the difference was not statistically significant (P=0.24). PBL with high-level MSI (cases 6 and 7) had mutations in the MMR genes: MLH1 and PMS2 or MSH6, respectively. Immunohistochemistry showed that these cases also had loss of MMR proteins (Table 2) as well as a high mutational burden. Other recur- rently-mutated genes included BRCC3, ATRX, FANCA, and BRIP1.
Mutations in the mitogen-activated protein kinase (MAPK) pathway genes were observed in six of 11 (55%) cases. Eight of nine mutations, occurring in KRAS, NRAS, HRAS, and BRAF, are well-known activating hotspot muta- tions (RAS codons 12, 13, and 61, and BRAF codons 469 and 601). Four cases harbored mutations in multiple genes, including one case with concomitant KRAS G13D, NRAS G12D, and BRAF G469V mutations.
Mutations in NOTCH signaling pathway genes were detected in five of 11 (45%) cases. Putative gain- or loss-of- function mutations in genes encoding the NOTCH family of proteins were the most frequent. SPEN, a negative regulator of NOTCH signaling, was mutated in three cases. Many of the NOTCH pathway mutations were in PBL with MMR defects and one case exhibited mutations in multiple NOTCH pathway members at different time points.
Janus kinase (JAK)/signal transducer and activator of tran- scription (STAT) signaling pathway genes, including STAT6 (n=3), STAT3 (n=2) and SOCS1 (n=1), were mutated in four of 11 (36%) cases.
Immune surveillance pathway genes were mutated in four of 11 (36%) cases; FAS was mutated in all four cases, all EBV– PBL, and in the monomorphic PTLD (DLBCL) preced- ing one PBL. One PBL harbored concurrent FAS and CD58 mutations.
The monomorphic PTLD (plasmacytoma), which har- bored a NOTCH1 mutation, acquired a BRIP1 mutation upon transformation. In contrast, some of the mutations
observed in the monomorphic PTLD (DLBCL), including a STAT3 variant, were not detected in the clonally related transformed PBL, suggesting divergent evolution from a common ancestor. No variants were identified in the poly- morphic PTLD preceding one PBL. The recurrent PBL showed both acquisition and loss of variants; the case with recurrent high-level MSI PBL (case 6) showed an increasing mutational burden over time.
Discussion
Our understanding of PT-PBL pathogenesis is limited and its molecular underpinnings are largely inferred from those reported for HIV-related PBL. Transcriptional analyses have revealedupregulationofJAK-STATpathwaygenesandsim- ilarities between PBL and plasma cell neoplasms (multiple myeloma [MM] and extra-osseous plasmacytoma) and dif- ferences between immunocompetent DLBCL and PBL, the latter displaying increased expression of MYC and MYB tar- get genes and genes regulating plasma cell differentiation and decreased expression of B-cell receptor and NF-kB sig- naling pathway genes.7,9 However, microRNA expression analysis has suggested two different subclasses of PBL, resembling either Burkitt lymphoma or extra-osseous plas- macytoma.10 Chang et al. documented overlapping chromo- somal aberrations between PBL and immunocompetent DLBCL as well as PBL-specific segmental gains at chromo- somes 1p and 1q by array comparative genomic hybridiza- tion.5 Whole exome sequencing has uncovered recurrent gains at 1q21, 6p22 and 11p13, loci that contain several genes (histones, IL6R, MCL1, and CD44) mechanistically linked with B-cell lymphomagenesis.7
MYC deregulation, due to rearrangement, amplification, or activation of certain signaling pathways (e.g., STAT3) is considered important for the development of PBL.11,17-19 The frequency of MYC abnormalities (rearrangement and/or gain) in our series (55%) was higher than that reported pre- viously for PT-PBL (38%), but within the range (36-69%) reported for PBL arising in other settings.2,3,7,11,12,17,19 In contrast to the study of mostly HIV-related PBL by Valera et al., MYC rearrangements were more common in EBV– PT-PBL,19 and in some cases, the rearrangement was detected upon PTLD transformation to PBL or at disease relapse.
Recently, whole exome sequencing and targeted sequenc- ing studies have unveiled the mutational landscape, primari- ly of HIV-related PBL,7,11 but until now the spectrum of genetic alterations underlying PT-PBL has not been explored.
204
haematologica | 2022; 107(1)