Page 158 - 2021_10-Haematologica-web
P. 158
J.E. Ramis-Zaldivar et al.
fiers (EP300, TET2, KMT2D, KMT2C, TAF1, ARID1A, HIST1H1D, HIST1H1E), cell cycle (TP53, MYC) and the NFκB pathway (CARD11, NFKBIE, TRAF3, MYD88) (P<0.05) (Figures 2E and 3; Online Supplementary Table S5). In terms of copy number alterations, EBV-negative PBL had higher levels of genetic complexity (16 vs. 9 alterations per case; Wilcoxon test, P=0.08) and recurrent 17p13 alterations (losses and CNN-LOH; Fisher test, P=0.025) (Figure 3). Differential gene expression analysis between EBV-positive (n=8) and EBV-negative (n=4) PBL identified seven differen- tially expressed genes (t-test, false discovery rate [FDR] <0.2 and fold change >±1) (Online Supplementary Table S3). Gene ontology analysis of those genes showed that EBV-negative cases had lower expression of genes related to hypoxia and p53 in the cardiovascular system (FDR=0.006) and p53 sig- naling pathways (FDR=0.13).
Clonal evolution in plasmablastic lymphomas
To analyze the hierarchy of genetic events in PBL, the cancer cell fraction was determined for 357 alterations (including 250 copy number alterations and 107 mutations) from 24 PBL cases, of which 31% were clonal (cancer cell fraction >85%) (Online Supplementary Tables S4 and S5). The
majority of alterations showed a wide spectrum of cancer cell fractions, except for TP53 mutations, 17 losses and 13q deletions which were mainly clonal (Shapiro test; P<0.05) (Online Supplementary Figure S7). Interestingly, clonal and subclonal mutations targeting recurrently mutated genes (STAT3, NRAS, TP53 and MYC) affected the same protein domains, suggesting that those subclonal driver mutations confer similar advantage to the cell (Online Supplementary Figure S8).
Comparison with overlapping lymphoma entities
Copy number data of 35 cases of BL20 were re-evaluated for comparison with the current PBL series (Online Supplementary Figure S9). PBL had higher levels of genomic complexity than BL (12.2 vs. 5.97 alterations/case; Wilcoxon test, P<0.01) with specific gains of 1q32.2-q44, 7p, 8q23.3, 11p13-p11.2, 11q23.3-q25, 12p13.32-p13.2, and 19p13.3- p13.12. We also compared the mutational landscape of our series with that of 28 BL.24 PBL displayed specific NRAS, STAT3 and EP300 mutations, fewer MYC mutations, and lack of aberrations in genes reported to be specifically asso- ciated with BL pathogenesis such as ID3, SMARCA4 and TCF3 (Fisher test; P<0.05) (Figure 4A).
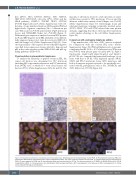
ABCD
EFGH
Figure 1. Plasmablastic lymphoma morphology and immunophenotype. Morphology and immunophenotype of case PBL6. (A) Plasmablastic lymphoma (PBL) with plasmacytic differentiation (hematoxilin-eosin, original magnification x400). The tumor cells were negative for (B) CD20, (C) showed weak and focal expression for PAX5, (D) were negative for CD56 and (E) also presented weak and focal positivity for CD79a. The case was positive for (F) EBER, (G) MUM1/IRF4 and (H) CD138 (magnification x400). (I, J) Fluorescence in situ hybridization (FISH) analysis identified a MYC rearrangement (I) but absence of BCL2 rearrangements (J). Yellow arrows indicate FISH signal constellations.
2686
haematologica | 2021; 106(10)