Page 148 - 2021_10-Haematologica-web
P. 148
A. Vogelsberg et al.
ABCD
E
F
G
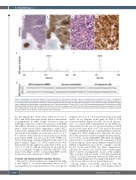
Figure 1. Morphology and molecular findings of the clonally related in situ follicular neoplasia and de novo high-grade B-cell lymphoma of case 1. (A) “Reactive” lymph node (LN) with in situ follicular neoplasia (ISFN) stained with hematoxylin and eosin (H&E). Note the intact lymphoid architecture. Inset: higher magnification of a strongly CD10 positive germinal center (GC). Original magnification 25x and 100x. (B) Strong BCL2 expression in GC colonized by ISFN. Inset: MIB1 stain with higher magnification demonstrating a low proliferation index. Original magnification 25x and 100x. (C and D) LN biopsy depicting high-grade B-cell lymphoma (HGBL) stained with H&E and BCL2. Original magnification 400x. (E and F) Immunoglobulin κ light chain (IGK) GeneScan analysis demonstrating a rearrangement involving the κ deleting element (Kde) with matching monoclonal peaks of 279 nucleotides (nt) in both lesions. (G) Sequencing of the BCL2 breakpoint revealed an identical BCL2/JH junction in both samples, confirming their clonal relationship.
the t(14;18)(q32;q21) translocation (Table 2). In case 5, BCL2 and IGH break-apart probes did not demonstrate re-arrangements in either sample. However, using an IGH/BCL2 dual-color, double fusion probe, both ISFN and DLBCL showed an aberrant hybridization pattern with a single fusion signal, suggesting a cryptic BCL2 translocation. Amplification of the BCL2 breakpoint was successful for the samples of seven cases (cases 1, 3, 6, 7, 8, 9, and 10), with six breaks located in the major break- point region (MBR) and one (case 8) in the 3’MBR sub- cluster. Sequencing confirmed identical breakpoints for all paired samples (Figure 1G). MYC translocations were demonstrated in the aggressive component of three cases (cases 1, 8, and 9), with an additional break in BCL6 in case 1, resulting in a diagnosis of HGBL with DH or TH, respectively (Figure 2).The corresponding ISFN and FL lesions showed no alterations of MYC or BCL6.
Clonality and immunoglobulin sequence analysis
The results of the IG analysis are summarized in Table 2 and Figure 2. A clonal relationship based on IG rearrangements was demonstrated for seven of ten paired
samples (cases 1, 2, 4, 7, 8, 9, and 10) by NGS of the IGH and/or by an identical clonal peak in IGH or IGK GeneScan analysis (Figure 1E and F). In case 10, the pres- ence of a clonal IGH rearrangement in the ISFN was demonstrated by the use of clone-specific primers, which produced the same peak of 127 base pairs in the paired ISFN, FL and DLBCL lesions, confirming their clonal rela- tionship (see Online Supplementary Appendix). In case 6, NGS demonstrated a clonal rearrangement in the DLBCL, but not in the corresponding ISFN, although both samples were shown to be clonally related by sequencing of their BCL2 breakpoint. Clone-specific primers designed for the DLBCL rearrangement also failed to amplify a specific product in the paired ISFN (see Online Supplementary Appendix). In contrast, cases 3 and 5 did not exhibit ampli- fiable clonal IG rearrangements in any of the samples. Thus, together with the results of the BCL2 breakpoint analysis, a clonal relationship between the ISFN and the corresponding lymphomas was firmly established for five of six de novo and four of four transformed cases.
Among samples successfully sequenced with the Lymphotrack Assay, we found novel N-glycosylation
2676
haematologica | 2021; 106(10)